Fig. 3.
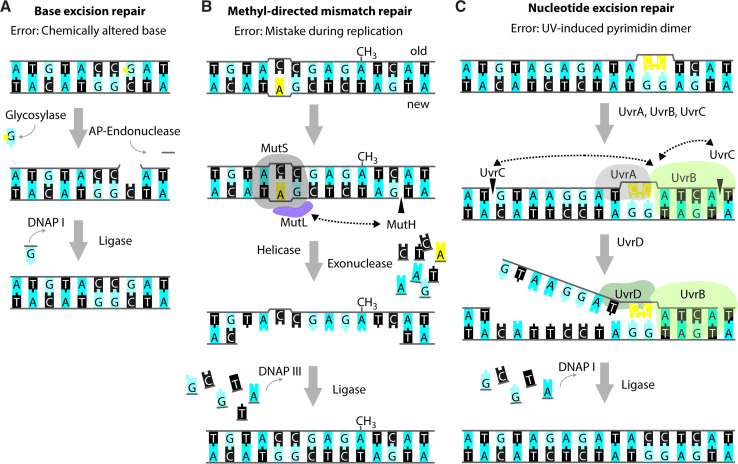
Repair of single-strand DNA lesions. Schematic representations of the modus operandi of base excision repair (BER) (a), methyl-directed mismatch repair (MMR) (b) and nucleotide excision repair (NER) (c). The lesions in the figure serve merely as examples as the aforementioned repair pathways are capable of repairing a variety of different lesions. In all examples, bases are shown as blocks using the one-letter code, the deoxyribose-phosphate moiety is depicted as a grey line. Incisions are indicated by black triangles penetrating the sugar–phosphate backbone. a Example of BER acting on a chemically altered base (denoted by the yellow star). The affected nucleotide is removed by the subsequent action of a glycosylase and an AP-endonuclease. DNAP I re-synthesizes the missing part of the DNA strand and DNA ligase closes the nick. b Example of MMR acting on a wrongly incorporated adenine (in yellow). MutS binds to the site of the distortion and subsequently recruits MutL and MutH. MutH incises the newly synthesized, non-methylated strand at the sequence GATC. Subsequently, a DNA helicase and exonuclease unwind and degrade part of the newly synthesized strand, including the non-matching nucleotide(s). DNAP III and DNA ligase fill in the missing sequence. c Example of NER acting on a pyrimidine dimer (in yellow). The UvrAB-complex binds to the site of the lesion and promotes incisions 3′ and 5′ from the lesion by UvrC. Subsequently, the UvrD-helicase promotes dissociation of the contained stretch of DNA. Also in NER, DNAP I re-synthesizes the missing part of the DNA strand, and DNA ligase closes the nick