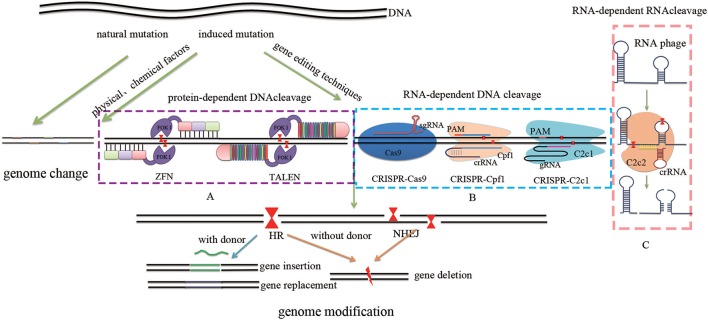
Figure 1.
Comparison of different GETs. Traditional methods include natural mutation via hybridization, induced mutation via ultraviolet light and x-ray (physical methods), as well as the use of benzene analogs and nitrous acid (chemical methods). These approaches offer a range of raw materials for evolution by randomly generating either autogenous or non-autogenous variation. Site-specific genome targeting technologies such as protein-dependent DNA cleavage systems (A) including ZFN and TALEN, as well as RNA-dependent DNA cleavage systems (B) including CRISPR-Cas9, CRISPR-Cpf1, and CRISPR-C2c1 can induce DSBs. In contrast, RNA-dependent RNA cleavage systems (C) such as SSB give rise to either random mutations via error-prone NHEJ or targeted mutations via error-free HR. These approaches achieve genome modification by inserting, deleting, or replacing targeted DNA sequences.