Figure 3. p53 and Sp1 bind competitively at the MMP-14 promoter.
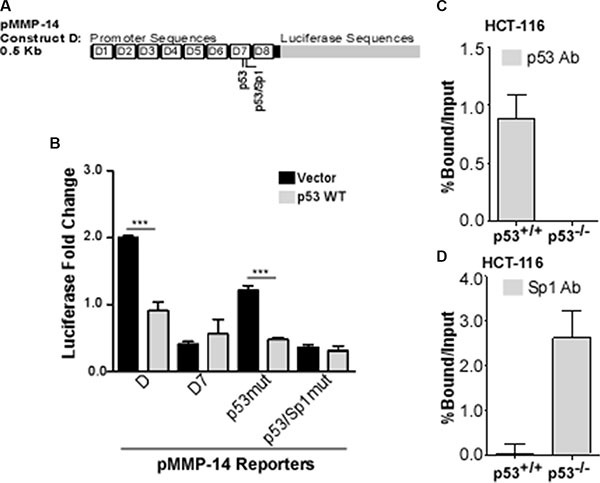
(A) Bioinformatic analysis identifies two potential p53 binding sites within deletion 7, the second of which is also a predicted Sp1 binding site. (B) Reporter gene assay was performed in COS-1 cells co-transfected with MMP-14 reporters for truncation D, D7, and mutated p53 or p53/Sp1 response elements (termed p53mut or p53/Sp1mut, respectively) along with either vector control or wild type p53 cDNAs followed by a luciferase assay. Renilla luciferase was used as a normalization control. Mutation at the second p53 binding element no longer responds to p53 expression. (C) ChIP-qPCR assay was performed using a p53 antibody to precipitate DNA from p53 wild-type (p53+/+) and p53 null (p53−/−) HCT-116 cells. Real time RT-PCR was then conducted using primers specific for MMP-14 promoter. MMP-14 promoter is precipitated by p53 antibody in HCT-116 (p53+/+) cells, whereas none is precipitated in p53−/− cells. (D) ChIP-qPCR was performed in HCT-116 cells with wild type or null p53 expression using anti-Sp1 antibody followed by a real time RT-PCR for MMP-14 promoter region. IgG and input samples were used as normalization controls. Results are expressed as percent of input. ***p < 0.001.
