Figure 2.
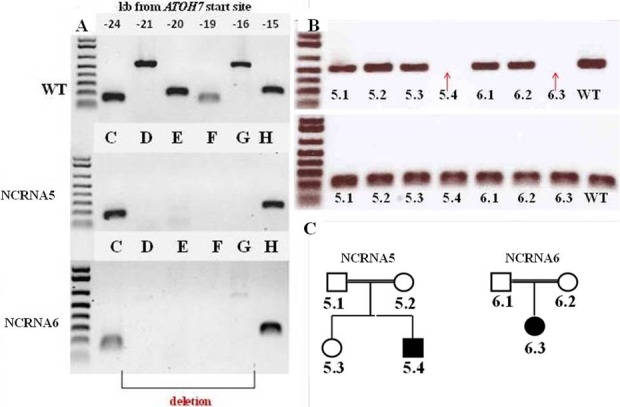
Polymerase chain reaction–based genotyping of homozygous 6523-kb deletion in 5′ATOH7 in NCRNA5 and NCRNA6 families. (A) A polymerase chain reaction–based deletion confirmation in the NCRNA5 and NCRNA6 families conducted as described by Ghiasvand et al.1 Letters (C, D, E, F, G, H) represent six adjacent amplicons in an NCRNA subject that amplify conserved noncoding elements ∼20 kb upstream of ATOH7 transcription start site. As it can be seen from the figure, C and H amplicons are present in both wild-type (WT) and mutant subjects, while the rest of the amplicons are missing in NCRNA5 and NCRNA6 family members. (B) Cosegregation results between family members using primers designed by Ghiasvand et al.1 In the first panel, primers for G and H amplicons are pooled, whereas in the second panel, only primers for H amplicons were used to verify if deletion of the area is present in the members of the family. Red arrows show deletion in affected family members compared with a WT control band. (C) Pedigrees of NCRNA5 and NCRNA6 families.
