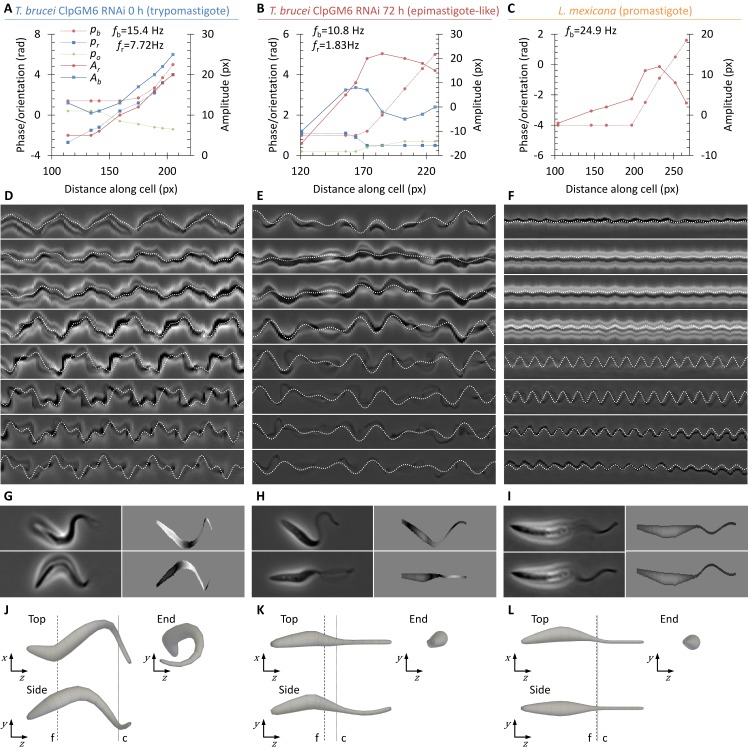
Fig 6. Determining the effective hydrodynamic shape of swimming promastigote, epimastigote-like and trypomastigote.
(A-C) Fitted parameters for cell body rotation phase (pr) and amplitude (Ar), flagellar beat phase (pb) and amplitude (Ab) and orientation of the beat plane (po) along the cell length of a representative (n = 8) trypomastigote uninduced ClpGM6 RNAi T. brucei (A), a representative (n = 7) epimastigote-like 72 h induced ClpGM6 RNAi T. brucei (B) and a promastigote L. mexicana (C) cell. Fitted cell rotation (fr) and flagellar beat (fb) frequencies are also indicated. (D-F) Example kymographs from the source high frame rate videomicrographs at different points along the cell overlaid with the movement trace from the fitted model parameters shown in (A-C). (G-I) Example still frames from the source high frame rate videomicrographs and the corresponding simulated cell appearance from the fitted model parameters shown in (A-C). Inferred height data is plotted through simulated phase defocus. (J-L) Three orthogonal views of the inferred three dimensional resting cell shape of a representative trypomastigote T. brucei (J), epimastigote-like T. brucei (K) and promastigote L. mexicana (L) cell. Resting cell shape was generated from the fitted model parameters shown in (A-C) by setting flagellum beat amplitude to zero, then rendering in three dimensions with cell body widths derived from the source high frame rate videomicrographs. Dashed lines indicate the approximate start of the flagellum (f) and end of the cell body (c).