Abstract
Cell-free cytoplasmic extracts prepared from Xenopus eggs and embryos have for decades provided a biochemical system with which to interrogate complex cell biological processes in vitro. Recently, the application of microfabrication and microfluidic strategies in biology has narrowed the gap between in vitro and in vivo studies by enabling formation of cell-size compartments containing functional cytoplasm. These approaches provide numerous advantages over traditional biochemical experiments performed in a test tube. Most notably, the cell-free cytoplasm is confined using a two- or three-dimensional boundary, which mimics the natural configuration of a cell. This strategy enables characterization of the spatial organization of a cell, and the role that boundaries play in regulating intracellular assembly and function. In this review we describe the marriage of Xenopus cell-free cytoplasm and confinement technologies to generate synthetic cell-like systems, the recent biological insights they have enabled, and the promise they hold for future scientific discovery.
Keywords: Encapsulation, Xenopus Egg Extract, Cellular Reconstitution, Microfluidics, Synthetic Cell, Compartmentalization
For over one hundred years, Xenopus has been used as a model system to investigate organismal development and to characterize fundamental processes that regulate cell structure and function. Xenopus is unique among model organisms in that its eggs have broad utility for studies both in vivo and in vitro. When fertilized, eggs facilitate in vivo studies on the mechanisms guiding embryogenesis, whereas cytoplasm isolated from its unfertilized eggs enables biochemical dissection of cell biological processes in vitro. Recently, Xenopus has also emerged as an important model organism for the study of human diseases; it has been particularly useful for identifying developmental mechanisms that are perturbed by genes mutated in patients (Duncan and Khokha, 2016). In this review, we highlight a new research area, cellular reconstitution, that takes advantage of Xenopus cytoplasmic extracts and confinement technologies to investigate how boundaries shape the spatial organization and activities of a cell.
Utility of Xenopus cytoplasmic extracts to dissect cell biological processes in vitro
Xenopus cell-free extracts are the cytoplasmic fraction that is isolated from intact eggs using centrifugation. This undiluted cytoplasm contains the protein milieu, organelles and subcellular structures present in an intact cell, and is competent to carry out many processes associated with the cell cycle in vitro (Desai et al., 1998; Hannak and Heald, 2006; Murray, 1991). Although studies in extracts are often complemented by experiments performed in vivo, the extract system provides a number of unique advances over studies in tissue culture. Importantly, cytoplasmic extracts are more tolerant of manipulations to the levels of essential cellular proteins, enabling a more comprehensive analysis of vital cell biological processes. In contrast, studies of core cellular processes in live cells are difficult because knockdown of an essential gene often causes cell cycle arrest or cell death. Additionally, the cell cycle state of Xenopus egg extracts can be readily synchronized, which enables individual parameters to be varied in isolation of other confounding factors present in growing cells. The system also lends itself to molecular investigation – the extracts are biochemically accessible and the availability of milliliter quantities of pure cytoplasm provides a means of fractionating and identifying the proteins that drive fundamental cellular activities.
For more than three decades, studies using cell-free extracts isolated from Xenopus eggs have produced groundbreaking molecular insights on the cell cycle control system, DNA replication and repair, chromosome condensation, transcriptional regulation, and regulated assembly or disassembly of cytoskeleton, kinetochores, organelles and other subcellular structures. Early studies focused on DNA replication and chromosome condensation in Xenopus extracts (Blow and Laskey, 1986; Hutchison et al., 1987; Lohka and Maller, 1985), guided by initial work using cell-free preparations from the frog, Rana pipiens (Lohka and Masui, 1983, 1984). The use of Xenopus egg extracts has facilitated a multitude of seminal discoveries including cyclin-based regulation of the cell cycle (Murray and Kirschner, 1989), and centrosome replication (Hinchcliffe et al., 1999), characterization of the DNA damage response pathway and cell cycle checkpoints (Cupello et al., 2016; Guo et al., 2000) and microtubule branching (Petry et al., 2013). Furthermore, fractionation and purification of proteins from egg extracts has led to the identification and characterization of proteins such as microtubule regulators, XMAP215, XKCM1 and TPX2 (Shirasu-Hiza et al., 2003; Tournebize et al., 2000; Walczak et al., 1996), and the actin nucleator Arp2/3 (Welch et al., 1997). These studies have provided key insights on the regulation of cytoskeletal dynamics. Additionally, Xenopus extracts maintain preeminence as a vertebrate model system for building functional subcellular structures in vitro; most notably assembly of the metaphase spindle and interphase nucleus (Desai et al., 1998; Heald et al., 1996; Kapoor et al., 2000; Merdes et al., 1996; Murray, 1991). This has led to important discoveries, such as the characterization of the bifunction role for the Ran GTPase in interphase nuclear transport and assembly (Zhang and Clarke, 2000) and as a controller of chromatin-mediated microtubule nucleation (Kalab et al., 2002; Ohba et al., 1999; Tsai et al., 2003).
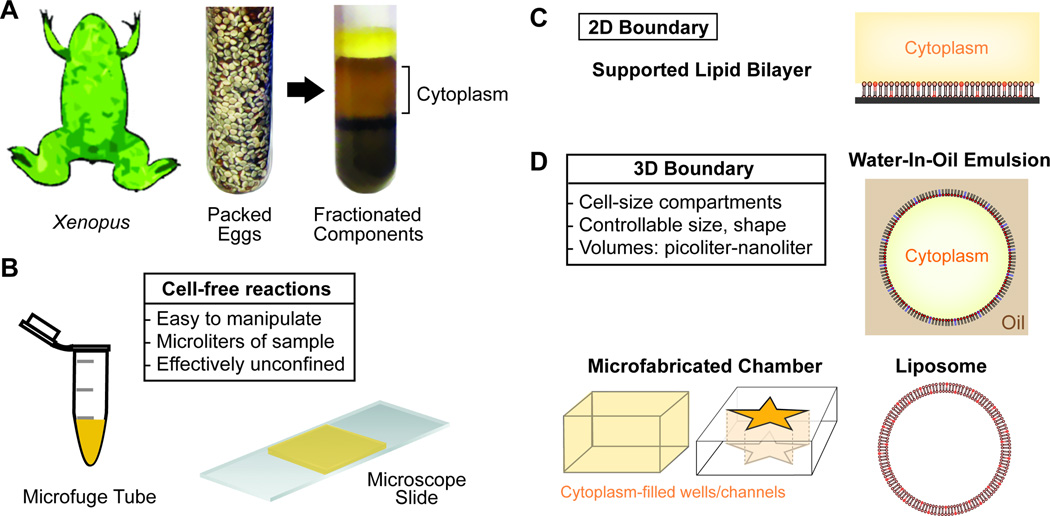
The preparation of cytoplasmic extracts from eggs of Xenopus laevis has been described thoroughly in previous work (Desai et al., 1998; Good, 2016; Hannak and Heald, 2006; Murray, 1991). Briefly, ovulated female frogs provide a source of thousands of unfertilized eggs which can be packed, crushed, and fractionated using centrifugation (Fig. 1A). Typically, one milliliter of the middle cytoplasmic layer is collected and supplemented with protease inhibitors and an energy regeneration mixture. The large volume of extract enables many reactions to be carried out in parallel and provides ample material for biochemical manipulation. Cytological reactions are often performed in a standard microfuge tube or imaged live on a microscope slide (Fig. 1B).
Figure 1. Xenopus Cell-free Egg Cytoplasmic Extracts and Strategies for 2D Confinement or 3D Encapsulation.
A. Eggs collected from the frog, Xenopus laevis, are fractionated to generate cell-free cytoplasm
B. The cytoplasmic extract is capable of carrying out complex cell biological processes in vitro in the absence of cell boundaries. Microliter volumes of cytoplasm are often activated and imaged in test tubes and on microscope slides.
C. Configuration for confinement of cellular reactions to a supported lipid bilayer surrounded by cytoplasm. The process of interest is initiated at or signals to a two-dimensional membrane whose composition is controllable.
D. Encapsulation of cytoplasmic extract reactions within cell-like boundaries. These stiffness of these boundaries varies: PDMS wells (rigid), water-in-oil emulsion (intermediate, lipid monolayer), or liposomes (soft, lipid bilayers). Importantly, compartment dimensions can be specified to encapsulate cell-size volumes of material; from picoliters to nanoliters
Variants of the canonical egg extract system
In addition to the traditional crude extracts prepared from Xenopus laevis eggs, new cytoplasm systems have been described which expand the scope of cell biological and developmental processes that can be characterized in vitro. For example, high speed extracts (HSEs) are prepared by clarifying crude extracts using ultracentrifugation, and these extracts provide an ideal system to investigate chromosome assembly or disassembly (Maresca and Heald, 2006). Furthermore, cell-free extracts can be prepared from eggs of the frog Xenopus tropicalis (Brown et al., 2007). These extracts form subcellular structures that are smaller than those found in egg extracts of Xenopus laevis, and thus can be utilized to identify factors that control intracellular size scaling (Helmke and Heald, 2014; Levy and Heald, 2010; Loughlin et al., 2011). Importantly, extracts are no longer limited to the egg stage; cytoplasm from early blastula embryos at various stages of development has been shown to function in vitro (Good, 2016; Good et al., 2013; Wilbur and Heald, 2013; Wuhr et al., 2008). This embryo extract system enables characterization of a specific cell biological process at varying stages of early embryogenesis, and may provide insights on how developmental regulation impacts intracellular function.
An important consideration is whether Xenopus egg extracts provide unique benefits that cannot be achieved using other cell-free systems. Extracts from prokaryotes (e.g. E. coli) and eukaryotes (e.g. wheat germ), along with rabbit reticulocytes and mammalian cell extracts have been widely used for protein synthesis in vitro (Bernhard and Tozawa, 2013; Endo and Sawasaki, 2006; Zemella et al., 2015). However, the prokaryotic systems cannot be used for eukaryotic cell biological studies and yeast extracts are not suitable for most cytological imaging. Notably, yeast extracts have been successfully used to reconstitute protein transport across ER-Golgi (Baker et al., 1988) but cannot be used to build large cytoskeletal structures and organelles that closely mimic those found in vertebrate species. In short, none of these simple extract system have been broadly established for cell biological applications. Additionally, unlike Xenopus egg extracts, the preparation of cell extracts from cultured cells, including mammalian tissue culture, leads to dilution of the cytoplasm. Intriguingly, recent work demonstrated the preservation of cell cycle activity ex vivo in cytoplasm aspirated from a single Drosophila embryo (Telley et al., 2012). However, only a few nanoliters of cytoplasm can be collected from an embryo using this method, and the centrifugation of thousands of embryos does not produce an active, cycling cytoplasm. Therefore, the Drosophila embryo system is not suitable for most standard biochemical analyses. In summary, given the limitations of other systems, Xenopus egg extracts remain the gold-standard for characterizing most cell biological processes in vitro.
Limitations of ‘test tube biochemistry’
Although undiluted Xenopus cytoplasm closely mimics the chemical composition of the egg or embryonic blastomeres, it lacks the structure and spatial organization that define a living cell. For example, studies using Xenopus egg extracts are typically carried out in microfuge tubes, requiring tens of microliters of cytoplasm, whereas the typical somatic cell volume is often less than ten picoliters. This difference of six orders of magnitude in the amount of cytoplasmic material has the potential to confound the interpretation of cellular activities that are studied in vitro. Additionally, cells have a defined architecture, including a cell boundary – the plasma membrane – which dictates the volume and shape of cell, controls protein subcellular localization, and provides mechanical feedback to internal processes. These and other variables may lead to diverging results in cells and test tubes (Minton, 2006). Therefore, to fully characterize a cell biological processes in a more cell-like context it is necessary to confine or encapsulate Xenopus cytoplasmic extracts. This experimental strategy is often described as ‘cellular reconstitution’ (Liu and Fletcher, 2009). In the following sections we describe recent studies that explored how boundaries regulate subcellular processes and structures, including cytoskeletal architecture and dynamics, organelle growth, and intracellular signaling.
There are a variety of strategies for generating boundaries to confine extract reactions to a two-dimensional membrane or within a micron length scale compartments, a subset of which have been discussed previously (Vahey and Fletcher, 2014). Historically, studies seeking insights on the chemical and physical impacts of boundaries relied on forming lipid bilayers on top of a solid support (Mueller et al., 1962; Richter et al., 2006; Tamm and McConnell, 1985). Because supported bilayers can promote specific associations of proteins on the membrane surface, they can be used to assess interactions between intracellular components and the cell boundary. More recently, research on the impacts of cell boundaries has been extended by three-dimensional compartmentalization. This includes, confinement of protein solutions within microfabricated channels and chambers containing a rigid boundary, and encapsulation within cell-size emulsions or liposomes (Griffiths and Tawfik, 2006; Martino and deMello, 2016) whose lipid composition can mimic that of the plasma membrane. Variations on these techniques have applications for reconstituting complex cellular processes in vitro in a cell-like configuration, and provide a novel method for manipulating the microenvironment of fundamental cell biological processes.
2D confinement of Xenopus cytoplasmic reactions: Investigating subcellular assembly at the cell membrane
Liposomes and supported lipid bilayers (SLBs) can be used in conjunction with Xenopus cytoplasmic extracts to reconstitute cellular processes that normally localize to the plasma membrane. For example, actin-dependent structures, such as filapodia, are nucleated by components concentrated on the membrane. Additionally, many signaling proteins, including GTPases, contain an amphipathic helix, lipidated peptide sequence or lipid-interacting domain, that promotes their association with membranes. Thus, the presence of a 2D bilayer is a minimal requirement to recapitulate the spatial segregation of cytosolic and membrane-associated factors found in a cell.
Recently, it was shown that phosphoinositide containing SLBs promote the assembly of filapodia-like structures from the surface of the bilayer in the presence of Xenopus high speed extracts (Lee et al., 2010). The authors demonstrated that these dynamic structures were nucleated from Arp2/3 complexes containing N-WASP, which associated with PI(4,5)P2 in the membrane. Additionally, they demonstrated a temporal order of recruitment: the first wave consisted of Toca-1, N-WASP and Arp2/3, followed by second wave of actin and formins. More recently, this method has been extended to allow additional membrane formulations (Walrant et al., 2015), which should enable characterization of how membrane properties, such as fluidity and lipid clustering, influence actin polymerization.
The combination of supported bilayers and Xenopus egg extracts have also been used to characterize cytokinesis signaling from regions of overlapping antiparallel microtubules to the cell membrane (Nguyen et al., 2014). The authors assembled centrosome-like asters, which recruit components of cendralspindlin and the chromosome passenger complex, proximal to a phosphoinositide containing SLB. They demonstrated that, proximal to regions of overlapping microtubules, this assembly was sufficient to induce the recruitment of activation of RhoA and additional cleavage furrow proteins. This system provides a modular framework with which to dissect the molecular activities and spatial constraints required for cell division in vitro. The specialized requirements for stabilizing SLBs in the presence of Xenopus egg extracts have recently been described (Field et al., 2015; Nguyen et al., 2015).
These studies, which leverage a 2D boundary, provide unique insights that could not have been derived from bulk egg extracts in a test tube and represent a clear step toward the long-term goal of reconstituting an intact cell. However, although these studies utilize a bilayer composition that mimics the plasma membrane, they lack the three-dimensional constraints and limited cytoplasmic volumes present in a cell. In the next section, we describe techniques for 3D encapsulation.
3D encapsulation of cytoplasmic extracts in cell-like compartments
As with many previous advances in biology, the generation of micron-sized compartments required adoption of technological advances in the physical sciences. Specifically, encapsulation of cells and other biological components was facilitated by applying photolithography (Whitesides et al., 2001) to development of microfluidics technologies for generating tunably-sized emulsions (Garstecki et al., 2006; Link et al., 2004; Thorsen et al., 2001; Utada et al., 2005)
Initially, a number of important studies demonstrated the feasible of encapsulating purified proteins, including tubulin, and centrosomes in micropatterned compartments (Faivre-Moskalenko and Dogterom, 2002; Holy et al., 1997). This was further extended by adding motor proteins to the compartment boundary (Laan et al., 2012). More recently, microtubules and motor proteins, and actomyosin assemblies have been characterized inside cell-size emulsion droplets (Baumann and Surrey, 2014; Miyazaki et al., 2015; Sanchez et al., 2012). However these studies are limited because many complex cellular structures, including the centrosome, nucleus and mitotic spindle, cannot be reconstituted using purified components.
The first studies to encapsulate Xenopus egg extracts in cell-like compartments focused on the architecture and dynamics of compartmentalized cytoskeletal assemblies. For example, Jiminez and colleagues demonstrated formation of microtubule asters and actin filaments inside aqueous-in-oil emulsions droplets stabilized by a non-lipid surfactant (Jimenez et al., 2011). Subsequent experiments exploited Ran-mediated microtubule aster formation inside similar droplets (Hoffmann et al., 2013). In a separate study, the authors quantified actin flow inside emulsion droplets (Pinot et al., 2012), and demonstrated that a polymerized actin network can promote subcompartmentalization (Colin et al., 2016). Other researchers have confined Xenopus egg extracts in micropatterned wells or channels to characterize contraction of microtubule networks (Foster et al., 2015).
A major benefit of confinement is that it reveals the contribution of boundaries to regulation of subcellular activities. For example, the boundary can function as a passive organizing platform that dictates subcellular anchoring of specific proteins or activities. Additionally, it defines the physical dimensions of a cell, and thus volume, of the cytoplasmic material present. Further, it can act as a mechanical barrier that resists osmotic swelling and cytoskeletal growth. The following studies provided important new insights on cell biological processes by exploiting Xenopus egg extracts and 3D encapsulation techniques to overcome previous experimental limitations.
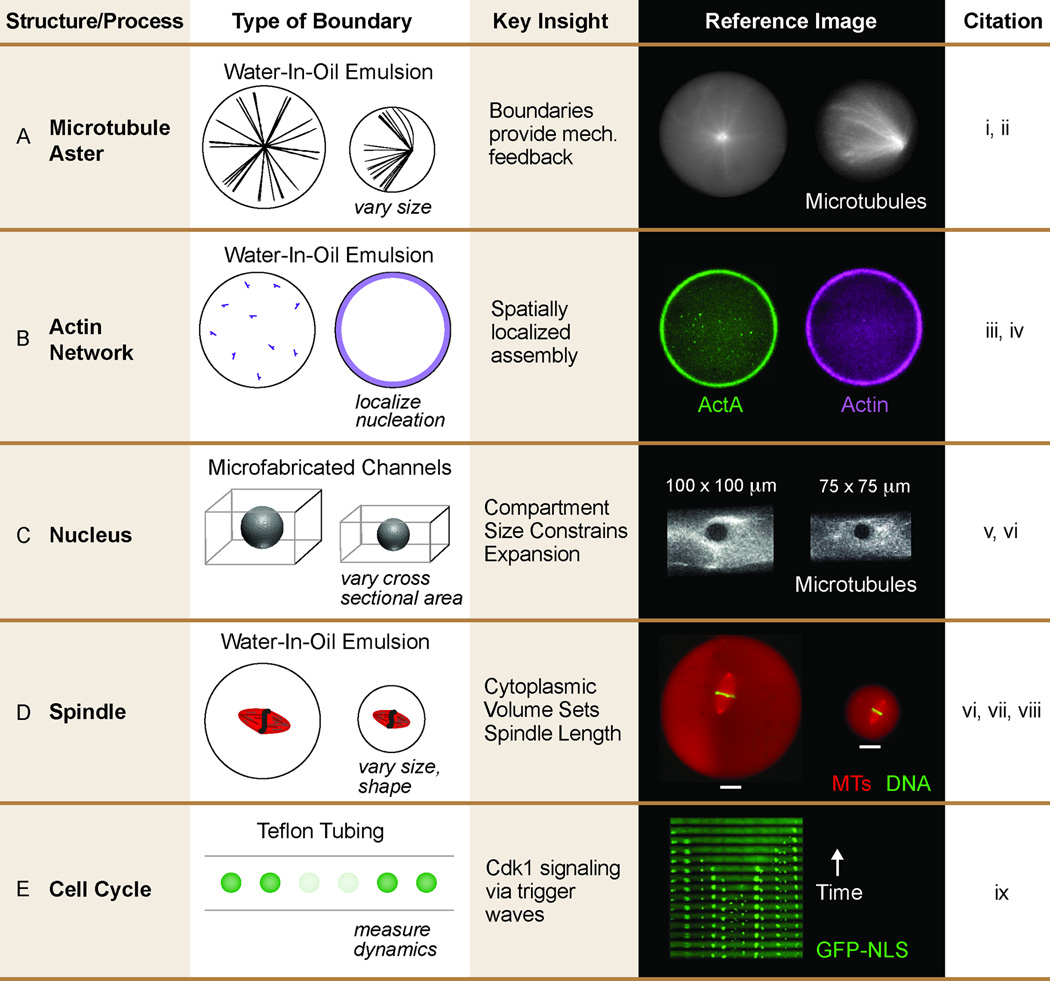
One prediction of confinement is that cytoskeletal polymers, assembled de novo, will experience a resisting force if the steady state size of the unencapsulated structure exceeds the dimensions of the compartment. Pinot and colleagues found that the size of the compartment – a cytoplasm-in-oil emulsion - impacted aster assembly; below a threshold size the lipid monolayer boundary mechanically restricted microtubule growth, causing buckling and reorganization of the canonical aster structure (Fig. 2A) (Pinot et al., 2009). This work is in agreement with previous studies using purified components that demonstrated that a boundary force will resist microtubule polymerization, leading to centering of an unanchored radial structure such as a microtubule aster (Faivre-Moskalenko and Dogterom, 2002; Holy et al., 1997).
Figure 2. Encapsulation of Xenopus Cytoplasmic Extracts in Microfabricated Chambers and Cell-Like Compartments.
A. Growth of radial microtubule asters is restricted by the droplet boundary in cytoplasm-filled water-in-oil emulsions. Below a threshold emulsion size, the microtubule growth causes the aster structure to buckle.
B. Spatial control of actin nucleation. Boundary-localized ActA initiates cortical actin polymerization in a cytoplasm-filled water-in-oil emulsions.
C. Nucleus growth scales with the cross-sectional area of the microchannel in which it is confined.
D. Metaphase spindle length is regulated by cytoplasmic volume. This scaling effect, in water-in-oil emulsions, is independent of compartment geometry.
E. Cell cycle duration is dependent on spatial position within cytoplasm-filled tubing.
Citations: i: Pinot et al, 2009, ii: Jiminez et al, 2011, iii: Shah et al, 2014, iv: Pinot et al, 2012, v: Hara et al, 2015, vi: Good et al, 2016, vii: Good et al, 2013, viii: Hazel et al, 2013, ix: Chang et al, 2013. Images courtesy of Z. Gueroui, K. Keren, C. Merten, J. Ferrell.
A cell boundary also provides a platform to spatially restrict subcellular assembly, including the cytoskeleton. For example, the cell cortex is composed of many proteins, including actin filaments that are nucleated by membrane-localized factors, such as the complex containing N-WASP and Arp2/3. The dynamic actin network that assembles near the plasma membrane is crucial for generating cell polarity, inducing cell motility and providing stiffness to the cell cortex. To investigate the spatial organization and biophysical properties of actin assembly, Shah and colleagues anchored the bacterial protein ActA to the boundary of emulsion droplets filled with Xenopus cytoplasmic egg extracts (Abu Shah and Keren, 2014; Abu Shah et al., 2015). When unanchored, ActA promoted actin polymerized throughout the droplet. However, when ActA was localized to the droplet boundary it nucleated an actin shell that resembles a minimal cell cortex (Fig. 2B). Intriguingly, the authors determined that this actin meshwork is dynamic, polarizes, and can generate force – features that mimic the properties of actin networks in cell.
Cell size is tightly regulated and cell volume varies only two-fold during the cell cycle of most symmetrically dividing cells grown in culture (Ginzberg et al., 2015). In contrast, cell size reduces dramatically during early embryo development, a period after egg fertilization in which cells divide without growing (Newport and Kirschner, 1982; Wuhr et al., 2008). A fundamental question is whether alterations in cell size also impact intracellular assembly or cellular function. A number of recent studies have begun to address this question, utilizing a cellular reconstitution approach that overcomes the challenges associated with manipulating cell size in vivo. This approach relies on the encapsulation of Xenopus cytoplasmic extracts in cell-size compartments, followed by analyses of how the sizes of intracellular structure vary as a function of compartment dimensions. As a first order approximation of cell boundaries, Hara and colleagues analyzed nucleus growth inside PDMS channels of varying cross-sectional area (Hara and Merten, 2015). They identified a minimal nuclear domain – a volume of material – below which nucleus growth rates began to slow. A closer approximation to a cell is a water-in-oil emulsion, with a spherical shape and lipid monolayer boundary can be specified to mimic cellular dimensions and the lipid composition of the cytoplasmic leaflet of the plasma membrane. This system, when coupled with Xenopus cytoplasm, allows variation of cell diameters from 10’s–100’s of microns, which mimics the size and composition of blastomeres present in blastula-stage embryos. Additionally, the shape of these emulsions can be controlled by compressing them or confining them in microfabricated channels or chambers of varying size (Good, 2016). An exciting recent discovery is the ability of the mitotic spindle, whose steady state size is relatively invariant in unencapsulated egg extracts (Brown et al., 2007), to sense and adapt to the size of the compartment in which it is assembled. Good and colleagues demonstrated that cytoplasmic volume sets metaphase spindle length in vitro, and this scaling closely matches the size scaling present in early embryo development (Good et al., 2013; Hazel et al., 2013). This result suggests that the physical dimensions of a cell regulate spindle size through a limiting pool of cytoplasmic components. These experiments utilized a passivated boundary rather than the full complement of plasma membrane lipids; therefore, these studies were carried out in the absence of a cell cortex. Thus, it remains possible that an interaction between astral microtubules and the cell cortex could further modulate spindle length, particularly during anaphase.
Theoretically, a cytoplasm-filled liposome surrounded by a fluid bilayer membrane would most closely resemble an intact cell. However, technological advancements are necessary to enable vesicle size control and improve the stability of these membranes, which are destabilized in the presence of Xenopus egg cytoplasm (unpublished data). One promising approach is the use of microjets for encapsulation. Pulsatile jetting of a solution will deform a planar lipid bilayer to generate a liquid-filled vesicle (Richmond et al., 2011; Stachowiak et al., 2009; Stachowiak et al., 2008). Microfluidic approaches for generating vesicle-like double emulsions may also succeed, although a concern is that residual oil or solvent present in the membrane limits their function as true bilayers. Additionally, formation of cytoplasm-filled vesicles using swelling or electroformation approaches is not feasible; the encapsulation efficiency is very low and osmolytes disrupt liposome formation. At present, microfluidic droplet formation provides the best combination of stability, size-control and throughput for generating cell-like compartments filled with undiluted Xenopus cytoplasmic extracts.
Not all intracellular processes scale with cell size. For example, in some organisms the timing of the cell cycle is invariant during early embryogenesis (Carvalho et al., 2009; Newport and Kirschner, 1982). However, a size-invariant process poses a conundrum for very large cells. As cell size increases, signals initiated from the lumen of a cell should take longer to diffuse to the cortex. Chang and colleagues investigated this problem by encapsulating cycling Xenopus egg extracts containing sperm nuclei in a Teflon tube (Chang and Ferrell, 2013). Normally, the period in which unencapsulated Xenopus extracts traverse the cell cycle is constant, averaging approximately thirty to forty minutes, tied to the expression or activation of cyclins and Cdks. However, when the cytoplasm is encapsulated in a tube, individual nuclei assemble and dissemble at distinct rates. This result is linked to spatial position within the tube because neighboring nuclei are more closely synchronized than distal nuclei. When nuclei in the tube are visualized as a kymograph, a wave behavior appears (Fig 2E), and these waves travel rapidly, at a rate that enables signaling from the centrosome to the cortex during the first cell division in the giant cells of the Xenopus embryo. The authors propose that these trigger waves result from interlinked positive and negative feedback loops regulating Ckd1 activation (Chang and Ferrell, 2013). Importantly, Cdk1 signals are transmitted more quickly than the rate of simple Brownian diffusion of activated Cdk1 between two distal sites.
Collectively, the encapsulation studies described in this section have generated exciting new insights on the biology of a cell. Importantly, these breakthroughs would not have been possible using unencapsulated egg extracts because they lack cell boundaries. Furthermore, many of the chemical and physical perturbations performed in these studies are not tolerated by live cells and therefore cannot be carried out in vivo.
Future Directions
I. Expanded toolkit for protein recruitment to compartment boundaries
To mimic a cell, cytoplasm must be encapsulated within compartments of appropriate size containing boundaries that at least partially mimic the lipid and protein composition of the plasma membrane. At present, the evolution of encapsulation technologies has far out-paced development of chemical strategies that are required to build a synthetic cell cortex and insert membrane proteins into a synthetic lipid bilayer. Alternatively, a simpler strategy is to recruit individual proteins to the head groups of lipids that comprise a compartment boundary, such as the lipid monolayer present in water-in-oil emulsions or the lipid bilayer present in liposomes (Stachowiak et al., 2012). Conceptually, this strategy would enable proper spatial positioning of peripheral membrane proteins; in the best case scenario, it could also be used to anchor the cytosolic domain of transmembrane proteins to the boundary, enabling formation of higher order cortical assemblies. However, achieving this goal requires the development of chemical tools that are compatible with cytoplasmic extracts and enable constitutive or inducible recruitment of proteins to a compartment boundary.
To date, synthetic lipid-protein interactions have been utilized to recruit tagged proteins to the surface of lipid monolayer or bilayer boundary through the incorporation of non-native lipids. For example, NTA-lipid has been widely used to induce recruitment of His-tagged proteins to one leaflet of liposomes and supported lipid bilayers (Gizeli and Glad, 2004; Stachowiak et al., 2012). Additionally, biotinylated lipids have been used to recruit oligomeric assemblies of streptavidin and biotinylated proteins to the boundary of water-in-oil emulsions (Vleugel et al., 2016). Covalent chemistry can be utilized to overcome weak protein-lipid interactions. Chemical couplers have also been paired with various classes of dendrimers (Percec et al., 2010; Turnbull and Stoddart, 2002), which can be used to form vesicle-like structures. However, the robustness of these strategies has only been established using purified proteins, not cytoplasmic extracts. Further testing is necessary to determine whether they will function in the context of a cytoplasm-filled, cell-like compartment. One localization mechanism that has proven compatible with Xenopus extracts is to label proteins with a strongly lipophilic dye, such as bodipy (Abu Shah et al., 2015). By doing so, it is possible non-specifically localize a protein to the water-oil interface of an emulsion droplet. However, we recommend caution with such an approach and suggest that orthogonal chemical dimerization strategies offer an optimal path forward.
For cells to function properly, proteins must be recruited to the plasma membranes at defined intervals of time and in specific spatial patterns. To recapitulate spatiotemporal regulation of protein localization inside cell-like compartments, light-inducible protein dimerization has emerged as the tool of choice. Recent studies have leveraged optogenetics to control the spatial localization of proteins and organelles with the cell (Ballister et al., 2014; Ballister et al., 2015; Guntas et al., 2015; Strickland et al., 2012; van Bergeijk et al., 2015), and induce protein assembly at the plasma membrane (Levskaya et al., 2009; Wagner and Glotzer, 2016). Additionally, there has been limited development of in vitro optogenetic tools (Hallett et al., 2016), which are expanding the types of manipulations that could be implemented for cellular reconstitution. We envision the inducible control of boundary-localization within an extract-filled compartment. This requires a configuration in which one-half of a light-inducible dimerization system is localized to the compartment boundary, while the protein of interest is fused to the other half of the optogenetic pair. In the best-case scenario, it would be possible to trigger recruitment of proteins to compartment boundaries or subcellular structures within a time scale of seconds. Additional practical considerations will likely have to be considered to achieve this goal.
II. Applications to Disease
The cellular reconstitution strategies described in this review provide a unique experimental framework to investigate molecular mechanisms that underlie human diseases, particularly those associated with improper sizing or spatial organization of a cell. For example, spindle positioning and chromosome segregation are critical for cell fate determination and organismal development. Intriguingly, mutations responsible for neurological diseases and some cancers are linked with defects in spindle orientation (Noatynska et al., 2012). Additionally, for a cell to function properly it must tightly regulate its dimensions and the sizes of its intracellular organelles (Ginzberg et al., 2015; Levy and Heald, 2012); deregulation of cell or nucleus size is associated with cellular senescence and diseases such as cancer (Li et al., 2015; Mitsui and Schneider, 1976; Sastre-Garau et al., 2004; Zink et al., 2004). To dissect mechanisms that contribute to these diseases it is necessary to control the dimensions and composition of the cell boundary; a feat that is not often possible in vivo. Therefore, investigating intracellular size control in cell-like compartment in vitro may provide paradigms for the contribution of organelle size misregulation to disease progression.
Xenopus cell-free cytoplasmic extracts are an emerging platform for screening small molecules that inhibit essential cellular pathways, including those involved in embryonic development and cancer progression (Broadus et al., 2015; Hang et al., 2012; Hardwick and Philpott, 2015; Sackton et al., 2014; Thorne et al., 2010). In contrast to screens in cell culture systems, Xenopus cytoplasm can be used to analyze the phenotypes of drugs whose uptake are restricted in live cells. Additionally, cell-free cytoplasm can be used to differentiate between effects that are broadly cytotoxic versus those that inhibit a specific cellular process. Intriguingly, extract encapsulation may be useful in combination with drug screening to identify molecules that affect nucleus size and shape (Hara and Merten, 2015), which are indicators of cancer progression (Zink et al., 2004).
Acknowledgments
We thank members of the Good lab for thoughtful comments and feedback. We also thank Peter Klein and Dan Kessler for their guidance and support of the Xenopus community at Penn. JGB is supported by a Cell and Molecular Biology Training Grant (NIH 5-T32-GM-007229-39). MCG is supported by grants from Burroughs Wellcome Fund, The March of Dimes Foundation, and Charles E. Kaufman Foundation.
References
- Abu Shah E, Keren K. Symmetry breaking in reconstituted actin cortices. Elife. 2014;3:e01433. doi: 10.7554/eLife.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Shah E, Malik-Garbi M, Keren K. Reconstitution of cortical actin networks within water-in-oil emulsions. Methods Cell Biol. 2015;128:287–301. doi: 10.1016/bs.mcb.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Ballister ER, Aonbangkhen C, Mayo AM, Lampson MA, Chenoweth DM. Localized light-induced protein dimerization in living cells using a photocaged dimerizer. Nature communications. 2014;5:5475. doi: 10.1038/ncomms6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Ayloo S, Chenoweth DM, Lampson MA, Holzbaur EL. Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Current biology : CB. 2015;25:R407–R408. doi: 10.1016/j.cub.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann H, Surrey T. Motor-mediated cortical versus astral microtubule organization in lipid-monolayered droplets. The Journal of biological chemistry. 2014;289:22524–22535. doi: 10.1074/jbc.M114.582015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard F, Tozawa Y. Cell-free expression--making a mark. Current opinion in structural biology. 2013;23:374–380. doi: 10.1016/j.sbi.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Broadus MR, Yew PR, Hann SR, Lee E. Small-molecule high-throughput screening utilizing Xenopus egg extract. Methods Mol Biol. 2015;1263:63–73. doi: 10.1007/978-1-4939-2269-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. The Journal of cell biology. 2007;176:765–770. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Desai A, Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Chang JB, Ferrell JE, Jr Mitotic trigger waves and the spatial coordination of the Xenopus cell cycle. Nature. 2013;500:603–607. doi: 10.1038/nature12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin A, Bonnemay L, Gayrard C, Gautier J, Gueroui Z. Triggering signaling pathways using F-actin self-organization. Scientific reports. 2016;6:34657. doi: 10.1038/srep34657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupello S, Richardson C, Yan S. Cell-free Xenopus egg extracts for studying DNA damage response pathways. The International journal of developmental biology. 2016;60:229–236. doi: 10.1387/ijdb.160113sy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Murray A, Mitchison TJ, Walczak CE. Chapter 20 The Use of Xenopus Egg Extracts to Study Mitotic Spindle Assembly and Function in Vitro. 1998;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Duncan AR, Khokha MK. Xenopus as a model organism for birth defects-Congenital heart disease and heterotaxy. Semin Cell Dev Biol. 2016;51:73–79. doi: 10.1016/j.semcdb.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17:373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Faivre-Moskalenko C, Dogterom M. Dynamics of microtubule asters in microfabricated chambers: the role of catastrophes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16788–16793. doi: 10.1073/pnas.252407099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Groen AC, Nguyen PA, Mitchison TJ. Spindle-to-cortex communication in cleaving, polyspermic Xenopus eggs. Molecular biology of the cell. 2015;26:3628–3640. doi: 10.1091/mbc.E15-04-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PJ, Furthauer S, Shelley MJ, Needleman DJ. Active contraction of microtubule networks. Elife. 2015;4 doi: 10.7554/eLife.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab on a chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- Ginzberg MB, Kafri R, Kirschner M. Cell biology. On being the right (cell) size. Science. 2015;348:1245075. doi: 10.1126/science.1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizeli E, Glad J. Single-step formation of a biorecognition layer for assaying histidine-tagged proteins. Anal Chem. 2004;76:3995–4001. doi: 10.1021/ac034855g. [DOI] [PubMed] [Google Scholar]
- Good MC. Encapsulation of Xenopus Egg and Embryo Extract Spindle Assembly Reactions in Synthetic Cell-Like Compartments with Tunable Size. Methods Mol Biol. 2016;1413:87–108. doi: 10.1007/978-1-4939-3542-0_7. [DOI] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic Volume Modulates Spindle Size During Embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AD, Tawfik DS. Miniaturising the laboratory in emulsion droplets. Trends in biotechnology. 2006;24:395–402. doi: 10.1016/j.tibtech.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes & development. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. Correlating in Vitro and in Vivo Activities of Light-Inducible Dimers: A Cellular Optogenetics Guide. ACS Synth Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang BI, Thorne CA, Robbins DJ, Huppert SS, Lee LA, Lee E. Screening for small molecule inhibitors of embryonic pathways: sometimes you gotta crack a few eggs. Bioorg Med Chem. 2012;20:1869–1877. doi: 10.1016/j.bmc.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nature protocols. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- Hara Y, Merten CA. Dynein-Based Accumulation of Membranes Regulates Nuclear Expansion in Xenopus laevis Egg Extracts. Developmental cell. 2015;33:562–575. doi: 10.1016/j.devcel.2015.04.016. [DOI] [PubMed] [Google Scholar]
- Hardwick LJ, Philpott A. An oncologists friend: How Xenopus contributes to cancer research. Developmental biology. 2015;408:180–187. doi: 10.1016/j.ydbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Helmke KJ, Heald R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. The Journal of cell biology. 2014;206:385–393. doi: 10.1083/jcb.201401014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Mazari E, Lallet S, Le Borgne R, Marchi V, Gosse C, Gueroui Z. Spatiotemporal control of microtubule nucleation and assembly using magnetic nanoparticles. Nature nanotechnology. 2013;8:199–205. doi: 10.1038/nnano.2012.246. [DOI] [PubMed] [Google Scholar]
- Holy TE, Dogterom M, Yurke B, Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:6228–6231. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CJ, Cox R, Drepaul RS, Gomperts M, Ford CC. Periodic DNA synthesis in cell-free extracts of Xenopus eggs. The EMBO journal. 1987;6:2003–2010. doi: 10.1002/j.1460-2075.1987.tb02464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AM, Roche M, Pinot M, Panizza P, Courbin L, Gueroui Z. Towards high throughput production of artificial egg oocytes using microfluidics. Lab on a chip. 2011;11:429–434. doi: 10.1039/c0lc00046a. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. The Journal of cell biology. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Mechanisms of intracellular scaling. Annual review of cell and developmental biology. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- Link DR, Anna SL, Weitz DA, Stone HA. Geometrically mediated breakup of drops in microfluidic devices. Phys Rev Lett. 2004;92:054503. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- Liu AP, Fletcher DA. Biology under construction: in vitro reconstitution of cellular function. Nature reviews Molecular cell biology. 2009;10:644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Maller JL. Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. The Journal of cell biology. 1985;101:518–523. doi: 10.1083/jcb.101.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Roles of cytosol and cytoplasmic particles in nuclear envelope assembly and sperm pronuclear formation in cell-free preparations from amphibian eggs. The Journal of cell biology. 1984;98:1222–1230. doi: 10.1083/jcb.98.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147:1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Heald R. Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol Biol. 2006;322:459–474. doi: 10.1007/978-1-59745-000-3_33. [DOI] [PubMed] [Google Scholar]
- Martino C, deMello AJ. Droplet-based microfluidics for artificial cell generation: a brief review. Interface Focus. 2016;6:20160011. doi: 10.1098/rsfs.2016.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Minton AP. How can biochemical reactions within cells differ from those in test tubes? Journal of cell science. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Chiba M, Eguchi H, Ohki T, Ishiwata S. Cell-sized spherical confinement induces the spontaneous formation of contractile actomyosin rings in vitro. Nature cell biology. 2015;17:480–489. doi: 10.1038/ncb3142. [DOI] [PubMed] [Google Scholar]
- Mueller P, Rudin DO, Tien HT, Wescott WC. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- Murray AW. Chapter 30 Cell Cycle Extracts. 1991;36:581–605. [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A Major Developmental Transition in Early Xenopus Embryos I: Characterization and Timing of Cellular Changes At the Midblastula Stage. Cell. 1982:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Field CM, Groen AC, Mitchison TJ, Loose M. Using supported bilayers to study the spatiotemporal organization of membrane-bound proteins. Methods Cell Biol. 2015;128:223–241. doi: 10.1016/bs.mcb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Groen AC, Loose M, Ishihara K, Wuhr M, Field CM, Mitchison TJ. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 2014;346:244–247. doi: 10.1126/science.1256773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noatynska A, Gotta M, Meraldi P. Mitotic spindle (DIS)orientation and DISease: cause or consequence? The Journal of cell biology. 2012;199:1025–1035. doi: 10.1083/jcb.201209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Percec V, Wilson DA, Leowanawat P, Wilson CJ, Hughes AD, Kaucher MS, Hammer DA, Levine DH, Kim AJ, Bates FS, et al. Self-assembly of Janus dendrimers into uniform dendrimersomes and other complex architectures. Science. 2010;328:1009–1014. doi: 10.1126/science.1185547. [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M, Chesnel F, Kubiak JZ, Arnal I, Nedelec FJ, Gueroui Z. Effects of confinement on the self-organization of microtubules and motors. Current biology : CB. 2009;19:954–960. doi: 10.1016/j.cub.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Pinot M, Steiner V, Dehapiot B, Yoo BK, Chesnel F, Blanchoin L, Kervrann C, Gueroui Z. Confinement induces actin flow in a meiotic cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11705–11710. doi: 10.1073/pnas.1121583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond DL, Schmid EM, Martens S, Stachowiak JC, Liska N, Fletcher DA. Forming giant vesicles with controlled membrane composition, asymmetry, and contents. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9431–9436. doi: 10.1073/pnas.1016410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter RP, Berat R, Brisson AR. Formation of solid-supported lipid bilayers: an integrated view. Langmuir. 2006;22:3497–3505. doi: 10.1021/la052687c. [DOI] [PubMed] [Google Scholar]
- Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB, Meaders J, Pfaff KL, Sigoillot F, Yu H, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–649. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T, Chen DT, DeCamp SJ, Heymann M, Dogic Z. Spontaneous motion in hierarchically assembled active matter. Nature. 2012;491:431–434. doi: 10.1038/nature11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu-Hiza M, Coughlin P, Mitchison T. Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. The Journal of cell biology. 2003;161:349–358. doi: 10.1083/jcb.200211095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Richmond DL, Li TH, Brochard-Wyart F, Fletcher DA. Inkjet formation of unilamellar lipid vesicles for cell-like encapsulation. Lab on a chip. 2009;9:2003–2009. doi: 10.1039/b904984c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Richmond DL, Li TH, Liu AP, Parekh SH, Fletcher DA. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4697–4702. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nature cell biology. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nature methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK, McConnell HM. Supported phospholipid bilayers. Biophysical journal. 1985;47:105–113. doi: 10.1016/S0006-3495(85)83882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Gaspar I, Ephrussi A, Surrey T. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. The Journal of cell biology. 2012;197:887–895. doi: 10.1083/jcb.201204019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol. 2010;6:829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nature cell biology. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, Prigent C, Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nature cell biology. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- Turnbull WB, Stoddart JF. Design and synthesis of glycodendrimers. Journal of biotechnology. 2002;90:231–255. doi: 10.1016/s1389-0352(01)00062-9. [DOI] [PubMed] [Google Scholar]
- Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Monodisperse double emulsions generated from a microcapillary device. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- Vahey MD, Fletcher DA. The biology of boundary conditions: cellular reconstitution in one, two, and three dimensions. Current opinion in cell biology. 2014;26:60–68. doi: 10.1016/j.ceb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleugel M, Roth S, Groenendijk CF, Dogterom M. Reconstitution of Basic Mitotic Spindles in Spherical Emulsion Droplets. J Vis Exp. 2016 doi: 10.3791/54278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E, Glotzer M. Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. The Journal of cell biology. 2016;213:641–649. doi: 10.1083/jcb.201603025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walrant A, Saxton DS, Correia GP, Gallop JL. Triggering actin polymerization in Xenopus egg extracts from phosphoinositide-containing lipid bilayers. Methods Cell Biol. 2015;128:125–147. doi: 10.1016/bs.mcb.2015.01.020. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- Wilbur JD, Heald R. Mitotic spindle scaling during Xenopus development by kif2a and importin alpha. Elife. 2013;2:e00290. doi: 10.7554/eLife.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Current biology : CB. 2008;18:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemella A, Thoring L, Hoffmeister C, Kubick S. Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems. Chembiochem : a European journal of chemical biology. 2015;16:2420–2431. doi: 10.1002/cbic.201500340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nature reviews Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]