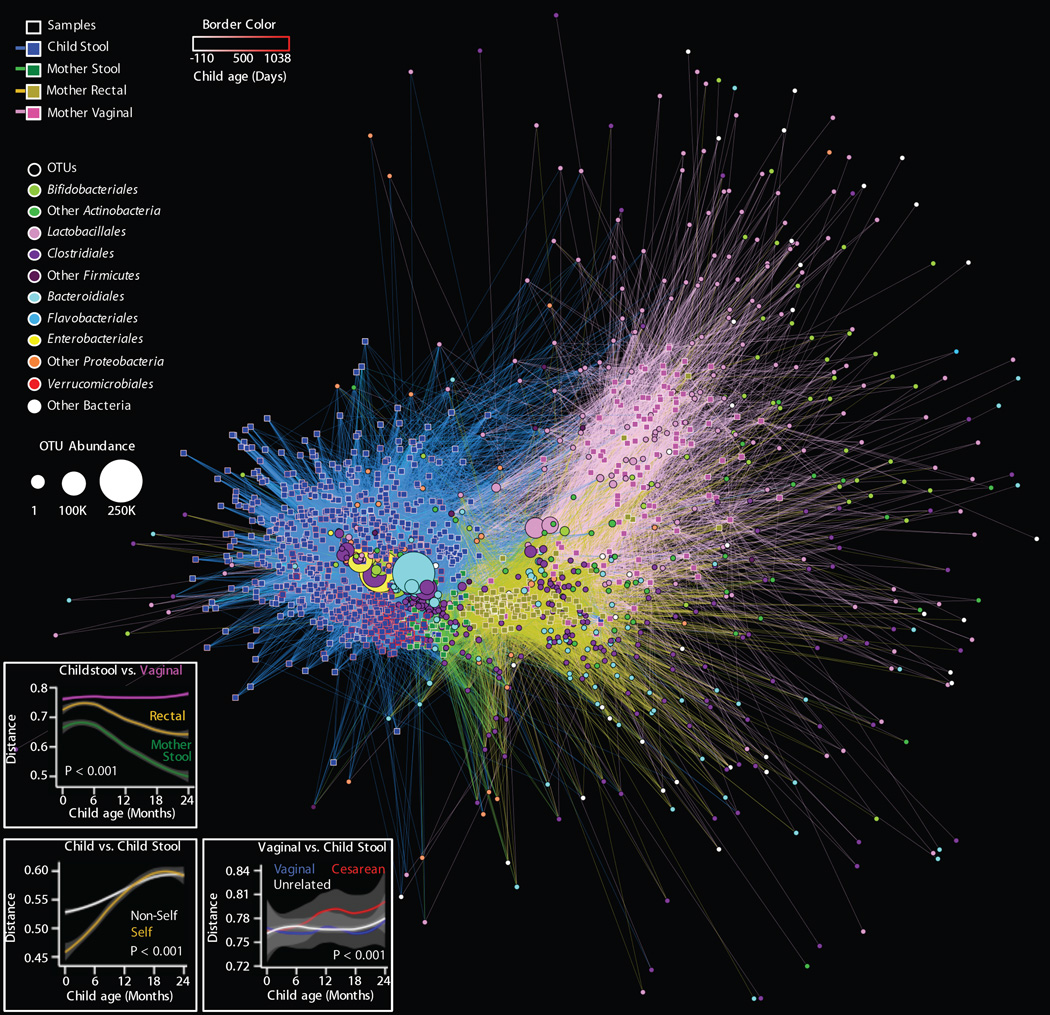
Fig. 6. Bipartite network comparing the relationships among all samples (squares) and OTUs (circles).
A, The distance between sample nodes and OTU nodes is a function of shared microbial composition. Samples with a large degree of OTU overlap (weighted by the number of observations of that OTU) form clusters. Edges connect a sample to each OTU detected in that sample, revealing shared OTUs between samples. Sample nodes and edges are colored by sample type; the border of sample nodes is a function of the age of the child, including pre-partum (negative) values for maternal samples (key at top-left). OTU nodes are colored by taxonomic family affiliation; the size of each OTU node is a function of that OTU's overall abundance, registered as OTU count in all samples (key at middle-left). See Fig 7 for specific analyses. B, Unweighted UniFrac distance between maternal vaginal, rectal, and stool microbiota and child stool microbiota as a function of child age. Shorter distance indicates greater similarity between microbial communities. C, Unweighted UniFrac distance between stool microbiota from the same child (self) and other children (non-self) as a function of the difference in age between sampling (Δ months). D, Unweighted UniFrac distance between maternal vaginal microbiota and stool microbiota of vaginally born dyads, unrelated children, or cesarean-delivered dyads as a function of child age. For panels B-D, lines indicate rolling-average mean values, grey shading = 95% CI. Grey shading = 95% CI. ANOVA P values are shown.