Fig. 3.
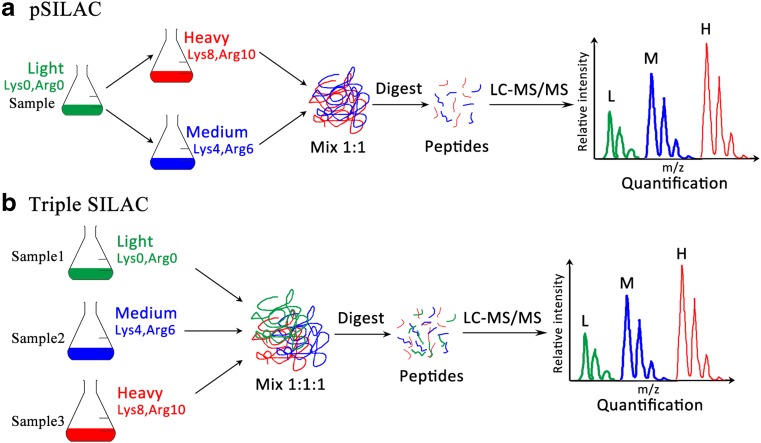
Stable isotope labeling with amino acids in cell culture (SILAC)-derived metabolic techniques. a pSILAC (pulsed SILAC) incorporated the stable isotope into proteins by adding “heavy” amino acids into the growth media. Cells are experimentally manipulated while growing in “light” (Lys0, Arg0) SILAC medium. Subsequently, the treated and control samples are transferred to distinctly labeled SILAC media, “heavy” (Lys8, Arg10) and “medium” (Lys4, Arg6). After one or a few doublings, samples are harvested and combined at the ratio 1:1. Proteins present before treatment will show up as a “light” peak (L) in the mass spectrograph and can be ignored. The effect of the treatment on protein production rates can be calculated as the ratio of signal at the “medium-heavy” (M) and “heavy” (H) peaks. b In triple SILAC, three samples can be analyzed at the same time, labeled with “light” (Lys0, Arg0), “medium” (Lys4, Arg6), and “heavy” (Lys8, Arg10) SILAC medium. Proteins were then combined and analyzed together by liquid chromatography tandem mass spectrometry (LC-MS/MS). In the MS spectra, each peptide appears as a triplet with distinct mass differences. The ratios between the samples are calculated directly by comparing the differences in the intensities of the peaks. “Lys0, Arg0”: unlabeled lysine and arginine; “Lys4, Arg6”: 2H4-lysine and 13C6-arginine; “Lys8, Arg10”: 13C6 15N2-L-lysine and 13C6 15N4-L-arginine