Abstract
Bisphenol A (BPA) is an endocrine‐disrupting chemical (EDC) prevalent in many household items. Rodent models and human epidemiological studies have linked this chemical to neurobehavior impairments. In California mice, developmental exposure to BPA results in sociosexual disorders at adulthood, including communication and biparental care deficits, behaviors that are primarily regulated by the hypothalamus. Thus, we sought to examine the transcriptomic profile in this brain region of juvenile male and female California mice offspring exposed from periconception through lactation to BPA or ethinyl estradiol (EE, estrogen present in birth control pills and considered a positive estrogen control for BPA studies). Two weeks prior to breeding, P0 females were fed a control diet, or this diet supplemented with 50 mg BPA/kg feed weight or 0.1 ppb EE, and continued on the diets through lactation. At weaning, brains from male and female offspring were collected, hypothalamic RNA isolated, and RNA‐seq analysis performed. Results indicate that BPA and EE groups clustered separately from controls with BPA and EE exposure leading to unique set of signature gene profiles. Kcnd3 was downregulated in the hypothalamus of BPA‐ and EE‐exposed females, whereas Tbl2, Topors, Kif3a, and Phactr2 were upregulated in these groups. Comparison of transcripts differentially expressed in BPA and EE groups revealed significant enrichment of gene ontology terms associated with microtubule‐based processes. Current results show that perinatal exposure to BPA or EE can result in several transcriptomic alterations, including those associated with microtubule functions, in the hypothalamus of California mice. It remains to be determined whether these genes mediate BPA‐induced behavioral disruptions.
Keywords: Brain, DOHaD, endocrine‐disrupting chemicals, estrogens, RNA‐seq
Introduction
Exposure to endocrine‐disrupting chemicals (EDCs), including bisphenol A (BPA), may increase the risk of neurobehavioral disorders typified by social impairments, such as autism spectrum disorders (Grandjean and Landrigan 2006; Landrigan 2010; Dietert et al. 2011; Miodovnik et al. 2011; Braun 2012; de Cock et al. 2012; Braun et al. 2014; Kalkbrenner et al. 2014; Kaur et al. 2014; Stein et al. 2015). Most EDCs are manufactured chemicals (Diamanti‐Kandarakis et al. 2009) with BPA being one of the most ubiquitous (He et al. 2009; Biedermann et al. 2010; Galloway et al. 2010). In 2013, production of this chemical was estimated to be approximately 15 billion pounds (GrandViewResearch, 2014). Its stability and pervasiveness (Environment Canada, 2008) has ensured continual exposure (Vandenberg et al. 2009). This chemical is detectable in the urine of 93% of the U.S. population (Calafat et al. 2008), as well as in fetal plasma, placenta (vom Saal et al. 2007), and breast milk (Vandenberg et al. 2007). In 2012, the FDA banned the production of baby bottles and sippy cups containing BPA https://www.federalregister.gov/documents/2012/07/17/2012-17366/indirect-food-additives-polymers. (accessed 23 January 2017). However, this restriction fails to address transfer of BPA across the placenta and through the milk (Ikezuki et al. 2002; Kawamoto et al. 2007; Balakrishnan et al. 2010; Nishikawa et al. 2010; Vandenberg et al. 2010). Moreover, fetuses and neonates lack many enzymes needed to metabolize BPA and may experience greater levels of active BPA than the mother (Ikezuki et al. 2002; Kawamoto et al. 2007; Nishikawa et al. 2010).
The fetus is especially vulnerable to endocrine disruption. Sex steroid hormones guide brain sexual differentiation throughout gestation and into the neonatal period (Arnold and Breedlove 1985; Adkins‐Regan 2005; McCarthy 2008). Environmental chemicals that mimic or interfere with sex hormone action may disturb this development (Palanza et al. 1999; Jasarevic et al. 2012; Kundakovic et al., 2013b). Broad ranges of behaviors are influenced by BPA exposure. Affected behaviors include reproductive, emotional, cognitive, and social behaviors and spatial reasoning (Berenbaum and Hines 1992; Mueller et al. 2008; Puts et al. 2008; Geary 2010). Many of the sociosexual deficits associated with BPA exposure in children and animals models are reviewed in Rosenfeld (2015). In our own studies, we tested two Peromyscus species, one that is polygynous and female uniparental (deer mice, P. maniculatus bairdii) and the other that is monogamous and biparental (California mice, Peromyscus californicus). In a mate choice experiment, females selectively reject deer mice males developmentally exposed to BPA (Jasarevic et al. 2011). Adult male California mice perinatally exposed to BPA show reduced territorial marking, a form of communication needed to protect the home range and mate from intruders, whereas exposed females demonstrate decreased exploratory and voluntary physical activity behaviors (Williams et al. 2013; Johnson et al. 2015c). Both male and female California mice developmentally exposed to BPA or ethinyl estradiol (EE, estrogen present in birth control pills) exhibit compromised parental care (Johnson et al. 2015b). These adult behavioral disruptions likely trace their origins to disturbances in normal brain programming during the perinatal period.
There is strong conservation in brain development and function across taxa, including in rodents and humans (Rice and Barone 2000; Howdeshell 2002). The hypothalamus is one of the primary brain areas governing many of these sociosexual behaviors. Thus, BPA might induce global transcriptomic changes in this brain region that is essential for guiding sociosexual behaviors. By using in situ hybridization, cDNA expression array, Northern blot, and qPCR approaches, prior rodent and zebrafish (Danio rerio) animal model, and in vitro cell culture studies report that BPA can alter individual candidate genes in hypothalamic regions or isolated neurons (Funabashi et al. 2001; Ceccarelli et al. 2007; Fukushima et al. 2007; Monje et al. 2007; Cao et al. 2012, 2013, 2014; Kundakovic et al., 2013a; Warita et al. 2013, 2014; Chen et al. 2014; Cano‐Nicolau et al. 2016). Collectively, these studies suggest BPA disrupts the hypothalamic expression of aromatase B (Cyp19a1b), nerve growth factor (Ngf), glucocorticoid receptor (Gr), estrogen receptor α (Esr1 and transcript variants), estrogen receptor β (Esr2), DNA methyltransferases (Dnmt1, 3a, 3b), methyl‐CpG‐binding protein 2 (Mecp2), kisspeptin (Kiss1), transforming growth factor‐β3 (Tgfb3), and progesterone receptor (Pr). A recent study suggests that in utero exposure of rats to BPA can induce select gene expression differences on postnatal day (PND) 1 (Arambula et al. 2016). However, to our knowledge, no previous studies have examined the comprehensive transcriptomic profile changes in the hypothalamus of a rodent model exposed to BPA or EE throughout the pre‐ and postnatal period, with the latter period approximating the third trimester of neural development in the hypothalamus, hippocampus, amygdala, and other brain regions in humans (Rice and Barone 2000; Howdeshell 2002). Such transcriptomic alterations may provide useful biomarkers of early exposure to these EDC. With this notion in mind, we used RNA‐seq to determine the global transcriptomic alterations in the hypothalamus of weanling (30 days of age) male and female California mice exposed to BPA throughout gestation and lactation. This time period was chosen as it represents the end of the exposure period to these chemicals and reflects the hypothalamic gene expression patterns prior to the observed adult‐onset behavioral deficits. The hypotheses at the outset were that (1) developmental exposure through the maternal diet to BPA or EE would induce global gene expression changes in the hypothalamus of juvenile California mice and (2) BPA and EE transcriptomic alterations would be dependent on offspring sex.
Materials and Methods
Animals and treatments
Founder out‐bred adult (60–90 days of age) California mice females and males were purchased from the Peromyscus Genetic Stock Center (PGSC) at the University of South Carolina (Columbia, SC), and placed in quarantine for a minimum of 8 weeks to ensure that they did not carry any common rodent pathogens. At the PGSC, P. californicus captive stocks have been bred to maintain their out‐bred status. We currently have our own breeding colony at the University of Missouri. As needed, additional California mice are purchased to maintain their out‐bred status. All experiments were approved by the University of Missouri Animal Care and Use Committee (Protocol #8693). Experiments were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Two weeks prior to breeding, virgin females, 8–12 weeks of age were randomly assigned to receive one of three diets: (1) a low phytoestrogen AIN 93G diet supplemented with 7% by weight corn oil to minimize potential phytoestrogenic contamination that would otherwise be present with inclusion of soybean oil in the diet, (2) the same diet supplemented with 50 mg BPA/kg feed weight, which we have documented to lead to internal serum concentrations close to those measured in pregnant women unknowingly exposed to this chemical (Jasarevic et al. 2011; Sieli et al. 2011), or (3) AIN93G diet supplemented with 0.1 parts per billion of EE, as the U.S. Food and Drug Administration (FDA) required estrogen‐positive control for BPA studies (vom Saal et al. 2005; Johnson et al. 2015a). The FDA has requested EE be included in BPA studies that may guide policy decisions based on the premise that BPA acts primarily as a weak estrogen (Vandenberg et al. 2009). The diets were started two weeks prior to breeding to span the periconceptional period. Females were maintained on these diets throughout gestation and lactation, as described previously (Jasarevic et al. 2011, 2013; Williams et al. 2013). To avoid any potential litter effects, only one male and one female offspring per litter were randomly chosen and examined in the current studies. At weaning, five replicates of each sex and group were euthanized, and brains were flash frozen on dry ice, and stored at −80°C until the hypothalamus was dissected, as described previously for mice (Kundakovic et al., 2013a). While no brain dissection guide for Peromyscus is available, the landmarks described in the Rat and Mouse Brain Dissection Guide Atlases (Paxinos and Franklin 2008; Paxinos 2013) can be used for California mice.
RNA isolation from hypothalamic samples
Hypothalamic RNA was isolated using Qiagen AllPrep DNA/RNA/miRNA Universal kit (Qiagen, Valencia, CA). Isolated DNA and miRNA will be used in future studies. The quantity and quality of the RNA was determined using a Nanodrop ND1000 spectrophotometer (Nanodrop Products, Wilmington, DE). The results were further confirmed by analyzing the RNA on the Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA). Only RNA with a RQN score above 8.0 was used for RNA‐seq analyses. Five different animals representing five different litters per each sex and each group were initially tested such that a total of 50 mice were initially screened. Similar numbers of replicates in other species have been successfully used to delineate transcriptomic changes following exposure to other environmental chemicals (Richter et al. 2014; Wood et al. 2014; Wirbisky et al. 2015; Arambula et al. 2016).
Illumina TruSeq RNA library preparation and sequencing
High‐throughput sequencing was performed at the University of Missouri DNA Core Facility. Libraries were constructed following the manufacturer's protocol with reagents supplied in Illumina's TruSeq Stranded mRNA Library Preparation kit. Briefly, the poly‐A containing mRNA is purified from total RNA (2 μg), RNA is fragmented, double‐stranded cDNA is generated from fragmented RNA, and the index containing adapters are ligated to fragment ends. PCR amplification was performed as follows: 98°C(0:30) + [98°C(0:10) + 60°C(0:30) + 72°C(0:30)] × 15 cycles + 72°C(5:00). The amplified cDNA construct were purified with Axyprep Mag PCR Clean‐up beads. Purified libraries were evaluated using the Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA), quantified with the Qubit 2.0 Flurometer (Invitrogen, Carlsbad, CA) using the quant‐iT HS dsDNA reagent kit, and diluted according to Illumina's standard sequencing protocol for sequencing on the HiSeq 2500 with a single end, 50 base read length. To maximize the number of reads per sample, only three samples were included in each lane, and the groups of three samples were randomized across treatments to avoid any confounding effects due to sequencing lane.
Gene expression analyses
Previously described methods (Givan et al. 2012) were modified to create a custom transcriptome and determine differential transcriptome expression. Specifically, 50‐mer RNA‐Seq reads from the Illumina HiSeq were first cleaned using scripts from the Fastx Toolkit ( https://github.com/agordon/fastx_toolkit) to trim the 3' ends of low‐quality (phred score < 20) bases and dropping reads when less than 50 bases were remaining. Reads were then filtered to exclude those that did not have a minimum of 95% of their bases with a quality score of 20 or more. Adapter sequence was removed with CutAdapt (Martin 2011) version 1.8.3. To create a final set of quality controlled RNA‐seq reads, foreign or undesirable reads were removed by sequence matching to the Phi‐X genome ( https://www.ncbi.nlm.nih.gov/genome/?term=NC_001422.1), the relevant ribosomal RNA genes as downloaded from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov) or repeat elements in RepBase version 20.02 (Jurka et al. 2005) using Bowtie version 1.1.1 (Langmead et al. 2009). Trinity version trinityrnaseq_r20140717 (Haas et al. 2013) was used to assemble the quality controlled reads into transcripts using default parameters. The final set of assembled transcripts is available for download ( https://genomics.ircf.missouri.edu/cgi-psd/ppsd.cgi?db=681). It is common to use paired‐end DNA sequence reads to de novo assemble transcriptomes. However, a priority was to balance the cost of the experiments with generating the data of most value. For this work, generating multiple replicates per sample of gene expression data was of higher value than generating a comprehensive transcript assembly. And it has been demonstrated that using single‐end (SE) reads for transcriptome assembly and quantification has relatively minor effects on gene‐level DE estimates, as described previously (Gonzalez and Joly 2013; Chhangawala et al. 2015). Thus, we chose to use SE data for this work. Functional annotation of the assembled transcripts was done by identifying proteins with significant sequence similarity within the NCBI NR sequence database using BLAST (Altschul et al. 1997).
To investigate the relationships between sample replicates, we first identified the list of genes that were differentially expressed (DE) between any two conditions of interest (DE genes). We collected the gene expression levels of all DE genes in each sample replicate. Upon visualizing the relationships between sample replicates based on the DE genes via a hierarchical cluster plot, samples from the EE group, EE.M1 and EE.M2, appeared to be significant outliers. To test the possibility that these samples were legitimate outliers and could be removed from the analysis, we first generated a distance matrix of the expression values of the DE genes between all sample replicates. The mean distance values were calculated for all sample replicates. Samples EE.M1 and EE.M2 had the highest mean distance from all other sample replicates. Upon removing these two values from the list of mean distances, the remaining distribution of values satisfied the Anderson–Darling test for normality, P = 0.06, as implemented in the nortest package in R ( https://cran.r-project.org/web/packages/nortest/index.html). Subsequently, the distance values for EE.M1 and EE.M2 were determined to be outliers using the Grubbs test for two outliers, P = 2.2e−16, as implemented in the outliers package in R ( https://cran.r-project.org/web/packages/outliers/index.html). Therefore, samples EE. M1 and EE.M2 were removed from all subsequent analyses discussed below. Subsequent to the removal of EE.M1 and EE.M2, no additional outliers were detected using the Grubbs test. Additionally, results from one of the EE female replicate samples were discarded due to 3X lower yield during the library generation. Lower yield may be due to a variety of reasons, but the potential for bias is high, and thus, this sample was not considered in subsequent analyses.
Differential expression of the transcripts in the remaining replicates was determined using RSEM version 1.2.15 (Li and Dewey 2011) and EdgeR version 3.8.2 (Robinson et al. 2010), as implemented in TrinityRNAseq. A transcript was considered differentially expressed between two conditions if the FDR value associated with the expression ratio was <0.05. Visualization of the gene expression results via the 3D plot of the principal component analysis was done using ggplot2 version 2.1.0 (Springer‐Verlag, 2009) and rgl version 0.96.0 (Adler 2016). Visualization of the differential expression via the heatmap was done using the ComplexHeatmap (Gu et al. 2016) package. Functional assessments of sets of differentially expressed genes such as pathway analyses and enrichment analyses were done using the TargetMine website (Chen et al. 2016). Before doing functional analyses, de novo‐assembled transcripts were mapped to human proteins based on extended amino acid similarity.
Results
General characterizations
The goal was to generate ~50 million reads per sample by loading three samples to each lane. The average number of raw reads was 59,965,113 and mapped reads was 43,462,565. Table 1 summarizes the alignment of RNA‐seq reads for each individual sample to the P. californicus genome sequence. For all three groups (BPA, EE, and controls) and both sexes, similar results were obtained in all of the categories. This number of reads and gene annotations is considered more than sufficient for a eukaryotic genome (Givan et al. 2012).
Table 1.
Summary of California mice hypothalamic RNA‐seq data generated and aligned to the reference genome sequence
| Sample type | Sample | Raw reads | QC | Mapped reads | Mapping efficiency |
|---|---|---|---|---|---|
| Control ♀ | 1 | 69,130,197 | 55,971,358 | 51,865,339 | 92.7 |
| 2 | 59,953,540 | 46,892,283 | 43,166,939 | 92.1 | |
| 3 | 63,537,487 | 49,756,404 | 46,269,276 | 93 | |
| 4 | 56,255,792 | 43,270,695 | 40,186,046 | 92.9 | |
| 5 | 70,576,520 | 54,790,689 | 50,805,216 | 92.7 | |
| Control ♂ | 1 | 46,376,849 | 34,835,622 | 33,238,813 | 95.4 |
| 2 | 65,673,113 | 51,651,522 | 47,823,627 | 92.6 | |
| 3 | 52,258,679 | 42,073,456 | 38,938,021 | 92.6 | |
| 4 | 59,220,633 | 47,041,870 | 43,660,840 | 92.8 | |
| 5 | 39,935,256 | 31,793,528 | 29,287,635 | 92.1 | |
| BPA ♀ | 1 | 45,592,976 | 36,777,665 | 33,894,109 | 92.2 |
| 2 | 62,081,241 | 48,332,836 | 44,626,692 | 92.3 | |
| 3 | 48,640,791 | 37,908,444 | 34,912,852 | 92.1 | |
| 4 | 64,901,425 | 49,432,382 | 45,486,404 | 92 | |
| 5 | 71,890,086 | 56,217,073 | 51,563,272 | 91.7 | |
| BPA ♂ | 1 | 66,308,474 | 49,724,724 | 45,792,562 | 92.1 |
| 2 | 56,296,984 | 44,408,114 | 40,927,183 | 92.2 | |
| 3 | 62,267,149 | 49,653,807 | 45,765,399 | 92.2 | |
| 4 | 68,054,969 | 53,980,003 | 50,117,318 | 92.8 | |
| 5 | 80,868,525 | 63,580,564 | 58,320,110 | 91.7 | |
| EE ♀ | 1 | 55,370,002 | 43,910,820 | 40,531,812 | 92.3 |
| 2 | 65,056,368 | 50,734,415 | 46,898,849 | 92.4 | |
| 3 | 56,097,553 | 42,583,645 | 39,390,432 | 92.5 | |
| 4 | 54,602,416 | 42,752,931 | 39,532,155 | 92.5 | |
| EE ♂ | 1 | 60,772,693 | 48,331,755 | 44,729,572 | 92.6 |
| 2 | 51,359,489 | 40,459,368 | 37,291,216 | 92.2 | |
| 3 | 65,978,847 | 52,139,890 | 48,467,572 | 93 |
Gene expression, association, and pathway differences
The PCA plot based on FPKM values shows a nonrandom distribution of points associated with samples (PC1 24.6%, PC2 10.2%, PC3 9.7%, PERMANOVA = 2e−04 from 10,000 permutations; Fig. 1A). Groupings of points are relatively obvious for BPA‐treated males, BPA‐treated females, EE‐treated males, and control males, whereas control females and EE‐treated females are intermingled. Even though the EE female samples cluster with the control female samples, it does not necessarily mean that the effects of EE on the female hypothalamus transcriptome are minor as further detailed below. A heatmap based on the hierarchical clustering of expression ratios for the differentially expressed genes revealed four main clusters containing primarily samples of BPA treatment, EE treatment, control females, and control males (Fig. 1B). There was no clear separation based on sex in either the BPA‐ or EE‐treated samples.
Figure 1.
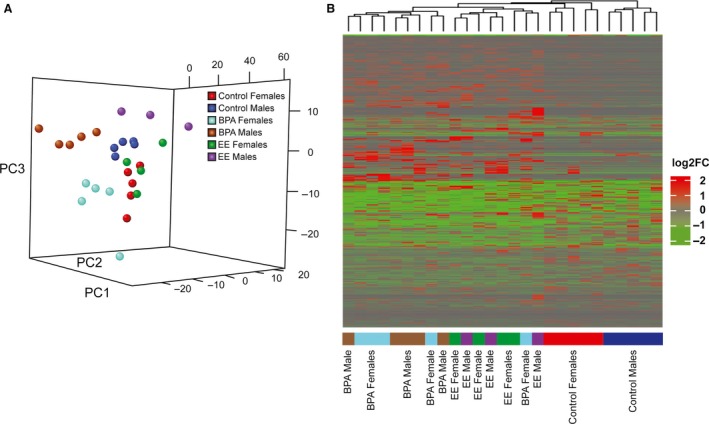
PCA plot and hierarchical heatmap. (A) PCA plot of the FPKM values of differentially expressed genes shows a nonrandom distribution of points (PERMANOVA = 2e−04 from 10,000 permutations), but control and EE females clustered together. (B) Hierarchical clustering and heatmap of expression ratios for the differentially expressed genes reveals primarily four groups: BPA‐treated, EE‐treated, control male, and control female groups.
Volcano plot comparisons revealed that our approach led to sufficient coverage to detect gene expression differences in BPA‐ or EE‐exposed males and females relative to their respective control groups (Fig. 2). Venn diagram comparison for females revealed select genes that were either increased or decreased in both BPA‐ and EE‐exposed individuals (Fig. 3A and B, Supporting Information File 1). However, each group possessed unique signature profile of genes that include transcripts that were increased and decreased in both groups. Likewise, BPA‐ and EE‐exposed males had select genes that were up‐ and downregulated in both groups (Fig. 3C and D, Supporting Information File 1), but more genes were either increased or decreased in one of the two treatment groups.
Figure 2.
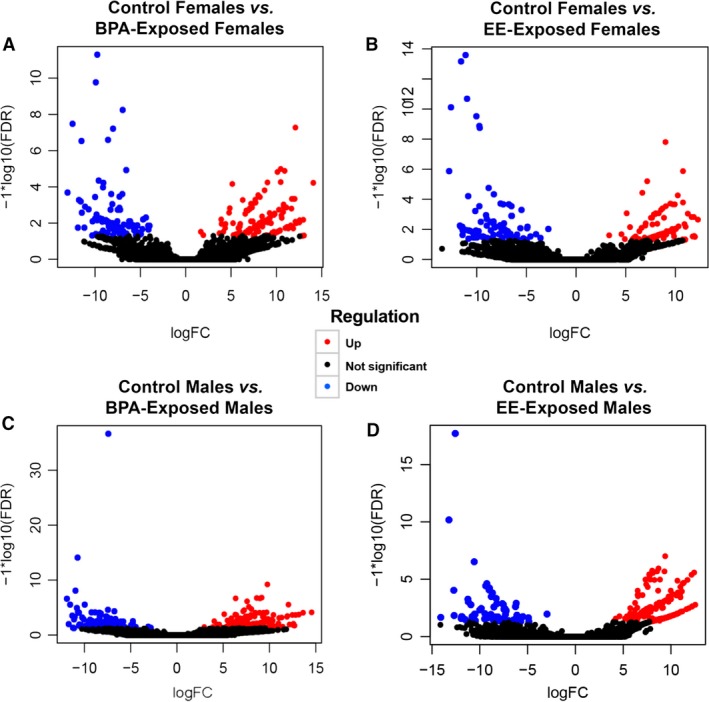
Volcano plots. (A) Control females versus BPA‐exposed females. (B) Control females versus EE‐exposed females. (C) Control males versus BPA‐exposed males. (D) Control males versus EE‐exposed males. Significantly downregulated genes in the treatment group versus controls are depicted in blue, black genes indicate that they are not significantly different, and genes delineated in red are increased in expression relative to controls.
Figure 3.
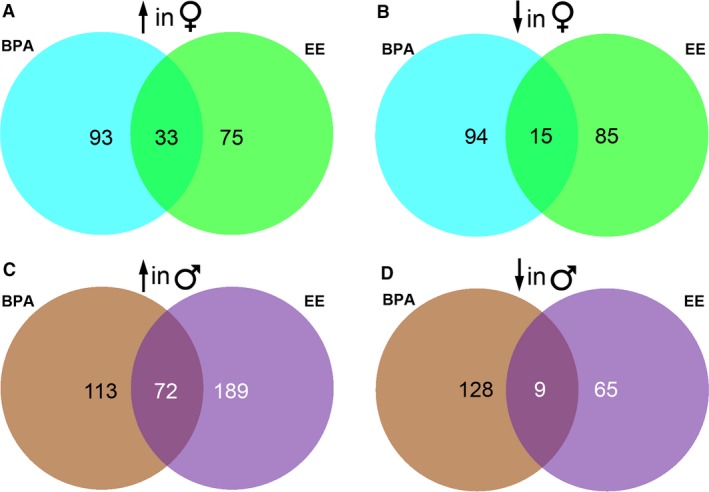
Venn diagrams. (A) Transcripts upregulated in BPA‐exposed females versus control females compared to those upregulated in EE‐exposed females versus control females. (B) Transcripts downregulated in BPA‐exposed females versus control females compared to those downregulated in EE‐exposed females versus control females. (C) Transcripts upregulated in BPA‐exposed males versus control males compared to those upregulated in EE‐exposed males versus control males. (D) Transcripts downregulated in BPA‐exposed males versus control males compared to those upregulated in EE‐exposed males versus control males.
As described earlier, EdgeR was used to identify the genes that differed in the following comparisons: control females versus control males, control females versus BPA females, control females versus EE females, control males versus BPA males, control males versus EE males, BPA females versus BPA males, and EE females versus EE males (Supporting Information File 2). Sex differences in hypothalamic gene expression between the three treatment groups are further addressed below. Tables 2 and 3 show the top 20 genes that are down‐ and upregulated, respectively, in BPA females versus control females. Tables 4 and 5 include the top 20 genes that are down‐ and upregulated, respectively, in EE females versus control females. Tables 6 and 7 show the top 20 genes that are down‐ and upregulated, respectively, in BPA males versus control males. Tables 8 and 9 include the top 20 genes that are down‐ and upregulated, respectively, in EE males versus control males. These collective tables indicate that each Treatment × Sex combination led to a unique signature set of genes. In other words, different genes were differentially regulated by BPA and EE and these depend on offspring sex. The exception is Kcnd3, which was downregulated in the hypothalamus of BPA‐ and EE‐exposed females relative to control females (bold in Tables 2 and 4). Tbl2, Topors, Kif13a, and Phactr2 were in the top 20 genes upregulated in BPA‐ and EE‐exposed females relative to control females (bold in Tables 3 and 5). No genes were shared in the top 20 genes differentially regulated in both BPA‐ and EE‐exposed males versus control males. Supporting Information File 3 provides gene differences between the three comparisons (BPA vs. control, EE vs. control, and BPA vs. EE) that were in common for both sexes.
Table 2.
Top 20 annotated genes downregulated in BPA‐exposed females compared to control females
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 9524 | TECR | Very long‐chain enoyl‐CoA reductase isoform 2 | 0.0002 | −13.043 |
| 114928 | GPRASP2 | Gprasp1 protein | 3.236E‐08 | −12.493 |
| 170506 | DHX36 | ATP‐dependent RNA helicase DHX36 | 0.0178 | −11.872 |
| 4594 | MUT | Methylmalonyl‐CoA mutase, mitochondrial | 0.0005 | −11.768 |
| 23683 | PRKD3 | Serine/threonine‐protein kinase D3 isoform 1 | 0.0006 | −11.574 |
| 90627 | STARD13 | stAR‐related lipid transfer protein 13 isoform 2 | 2.865E‐07 | −11.475 |
| 25828 | TXN2 | Thioredoxin, mitochondrial precursor | 0.0026 | −11.454 |
| 1741 | DLG3 | Disks large homolog 3 isoform 2 | 0.0171 | −11.179 |
| 3752 | KCND3 | Potassium voltage‐gated channel subfamily D member 3 isoform 1 precursor | 0.0013 | −11.092 |
| 65109 | UPF3B | Regulator of nonsense transcripts 3B isoform X2 (Peromyscus maniculatus bairdii) | 0.0017 | −10.696 |
| 23633 | KPNA6 | Importin subunit α‐7 | 0.0480 | −10.303 |
| 2342 | FNTB | Protein farnesyltransferase subunit beta isoform X2 (Macaca fascicularis) | 0.0079 | −10.281 |
| 8910 | SGCE | Epsilon‐sarcoglycan isoform X4 (Peromyscus maniculatus bairdii) | 0.0004 | −9.9968 |
| 54906 | FAM208B | protein FAM208B isoform X1 (Peromyscus maniculatus bairdii) | 0.0055 | −9.9953 |
| 9663 | LPIN2 | Mus musculus strain C57BL6/J chromosome 6 clone RP23‐258N2, complete sequence | 1.66E‐10 | −9.9061 |
| 501 | ALDH7A1 | α‐Aminoadipic semialdehyde dehydrogenase isoform X1 (Peromyscus maniculatus bairdii) | 0.0034 | −9.8387 |
| 287 | ANK2 | Ankyrin‐2 (Propithecus coquereli) | 0.0059 | −9.7829 |
| 55759 | WDR12 | Ribosome biogenesis protein WDR12 (Peromyscus maniculatus bairdii) | 0.0067 | −9.7639 |
| 4124 | MAN1A1 | Mannosyl‐oligosaccharide 1,2‐α‐mannosidase IA isoform X1 (Peromyscus maniculatus bairdii) | 0.0474 | −9.7301 |
| 81614 | NIPA2 | Magnesium transporter NIPA2 (Peromyscus maniculatus bairdii) | 0.0430 | −9.6497 |
Table 3.
Top 20 annotated genes upregulated in BPA‐exposed females compared to control females
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 26608 | TBL2 | Microtus ochrogaster guanine nucleotide binding protein (G protein), gamma 2 (Gng2), mRNA | 5.84E‐05 | 14.060 |
| 23008 | KLHDC10 | Kelch domain‐containing protein 10 | 0.0473 | 13.042 |
| 170506 | DHX36 | ATP‐dependent RNA helicase DHX36 isoform X2 | 0.0067 | 12.949 |
| 157378 | TMEM65 | Transmembrane protein 65 (Microtus ochrogaster) | 0.0071 | 12.571 |
| 10938 | EHD1 | EH‐domain containing 1, partial | 0.0089 | 12.542 |
| 3920 | LAMP2 | Lysosome‐associated membrane glycoprotein 2 isoform 2 precursor | 0.0092 | 12.409 |
| 4301 | MLLT4 | Afadin isoform X15 | 0.0102 | 12.165 |
| 9026 | HIP1R | Huntingtin‐interacting protein 1‐related protein isoform X1 (Mesocricetus auratus) | 0.0086 | 12.098 |
| 221937 | FOXK1 | Forkhead box protein K1 | 5.23E‐08 | 12.075 |
| 10210 | TOPORS | Mustela putorius furo UDP‐Gal:betaGlcNAc beta 1,4‐ galactosyltransferase, polypeptide 5 (B4GALT5), partial mRNA | 0.0148 | 11.784 |
| 63971 | KIF13A | Kinesin‐like protein KIF13A isoform X4 ( Peromyscus maniculatus bairdii ) | 0.0004 | 11.690 |
| 23274 | CLEC16A | Protein CLEC16A isoform X4 | 0.0015 | 11.641 |
| 129049 | SGSM1 | Small G protein signaling modulator 1 isoform X1 (Mesocricetus auratus) | 0.0133 | 11.569 |
| 9749 | PHACTR2 | Phosphatase and actin regulator 2 isoform X1 | 0.0160 | 11.394 |
| 55731 | FAM222B | Protein FAM222B isoform X1 (Peromyscus maniculatus bairdii) | 0.0161 | 11.172 |
| 253959 | RALGAPA1 | Ral GTPase‐activating protein subunit α‐1 isoform X4 | 0.018 | 10.986 |
| 8314 | BAP1 | Ubiquitin carboxyl‐terminal hydrolase BAP1 | 1.29E‐05 | 10.90 |
| 375 | ARF1 | Peromyscus maniculatus bairdii PHD finger protein 20‐like 1 (Phf20l1), transcript variant X3, mRNA | 0.0009 | 10.741 |
| 10401 | PIAS3 | E3 SUMO‐protein ligase PIAS3 (Microtus ochrogaster) | 0.001 | 10.780 |
| 26035 | GLCE | D‐glucuronyl C5‐epimerase (Peromyscus maniculatus bairdii) | 0.0009 | 10.557 |
| 113402 | SFT2D1 | Peromyscus maniculatus bairdii SFT2 domain containing 1 (Sft2d1), mRNA | 0.0051 | 10.592 |
Table 4.
Top 20 annotated genes downregulated in EE‐exposed females compared to control females
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 22864 | R3HDM2 | R3H domain‐containing protein 2 isoform X9 | 1.33E‐06 | −12.787 |
| 23017 | FAIM2 | Protein lifeguard 2 isoform 1 | 7.67E‐11 | −12.591 |
| 9671 | WSCD2 | WSC domain‐containing protein 2 | 0.0055 | −11.717 |
| 3149 | HMGB3 | High‐mobility group protein B3 isoform X2 | 6.54E‐14 | −11.604 |
| 5905 | RANGAP1 | Ran GTPase‐activating protein 1 isoform X3 | 0.0067 | −11.488 |
| 54982 | CLN6 | Peromyscus maniculatus bairdii CDP‐diacylglycerol‐inositol 3‐phosphatidyltransferase (Cdipt), transcript variant X2, mRNA | 0.0075 | −11.451 |
| 10390 | CEPT1 | Choline/ethanolaminephosphotransferase 1 isoform 1 | 0.0225 | −11.439 |
| 59338 | PLEKHA1 | Pleckstrin homology domain‐containing family A member 1 isoform X9 | 0.0109 | −11.437 |
| 9915 | ARNT2 | Aryl hydrocarbon receptor nuclear translocator 2 isoform X1 | 2.57E‐14 | −11.130 |
| 3752 | KCND3 | Potassium voltage‐gated channel subfamily D member 3 isoform 1 precursor | 0.0109 | −11.056 |
| 23378 | RRP8 | Ribosomal RNA‐processing protein 8 isoform 2 | 2.05E‐11 | −10.974 |
| 7690 | ZNF131 | Zinc finger protein 131, isoform CRA_a | 5.85E‐05 | −10.866 |
| 113829 | SLC35A4 | UDP‐sugar transporter protein SLC35A4 | 0.0412 | −10.820 |
| 140609 | NEK7 | Microtus ochrogaster NIMA‐related kinase 7 (Nek7), mRNA | 0.0116 | −10.817 |
| 10206 | TRIM13 | E3 ubiquitin‐protein ligase TRIM13 | 0.0126 | −10.684 |
| 745 | MYRF | Myelin regulatory factor isoform X6 | 0.0158 | −10.410 |
| 84915 | FAM222A | Protein FAM222A | 0.0154 | −10.371 |
| 4728 | NDUFS8 | NADH dehydrogenase (ubiquinone) iron‐sulfur protein 8, mitochondrial | 0.0198 | −10.334 |
| 2186 | BPTF | Nucleosome‐remodeling factor subunit BPTF isoform X6 | 0.0348 | −10.330 |
| 1973 | EIF4A1 | Eukaryotic initiation factor 4A‐I (Mesocricetus auratus) | 0.0348 | −10.289 |
Table 5.
Top 20 annotated genes upregulated in EE‐exposed females compared to control females
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 26608 | TBL2 | Microtus ochrogaster guanine nucleotide binding protein (G protein), gamma 2 (Gng2), mRNA | 0.0022 | 12.258 |
| 78986 | DUSP26 | Dual specificity protein phosphatase 26 | 0.0303 | 11.953 |
| 64400 | AKTIP | AKT‐interacting protein isoform 1 | 0.0271 | 11.904 |
| 5909 | RAP1GAP | Rap1 GTPase‐activating protein 1 isoform 2 | 0.0272 | 11.887 |
| 54467 | ANKIB1 | Peromyscus maniculatus bairdii RAD23 homolog B (Saccharomyces cerevisiae) (Rad23b), mRNA | 0.0290 | 11.738 |
| 63971 | KIF13A | Kinesin family member 13A isoform X4 | 0.0015 | 11.621 |
| 8266 | UBL4A | Peromyscus maniculatus bairdii ubiquitin‐like 4A (Ubl4a), mRNA | 0.0067 | 11.259 |
| 113178 | SCAMP4 | Secretory carrier membrane protein 4, isoform CRA_c | 0.0468 | 10.990 |
| 10210 | TOPORS | Mustela putorius furo UDP‐Gal:betaGlcNAc beta 1,4‐ galactosyltransferase, polypeptide 5 (B4GALT5), partial mRNA | 0.0472 | 10.973 |
| 10150 | MBNL2 | Peromyscus maniculatus bairdii muscleblind‐like splicing regulator 2 (Mbnl2), transcript variant X2, mRNA | 0.0044 | 10.917 |
| 9749 | PHACTR2 | Phosphatase and actin regulator 2 isoform D | 0.0490 | 10.900 |
| 4311 | MME | CD10 neutral endopeptidase 24.11 | 1.34E‐06 | 10.778 |
| 26035 | GLCE | d‐glucuronyl C5‐epimerase | 0.0002 | 10.774 |
| NA | CMPK1 | Cricetulus griseus cytidine monophosphate (UMP‐CMP) kinase 1, cytosolic (Cmpk1), mRNA | 0.044 | 10.735 |
| 100506658 | OCLN | Occludin, isoform CRA_b | 0.006 | 10.300 |
| 10659 | CELF2 | CUGBP Elav‐like family member 2 isoform 1 | 5.40E‐05 | 10.240 |
| 112970 | KTI12 | Protein KTI12 homolog | 0.0089 | 10.064 |
| 23114 | NFASC | Neurofascin isoform X12 | 0.0002 | 9.9501 |
| 2058 | EPRS | Bifunctional glutamate/proline–tRNA ligase isoform X1 | 0.0286 | 9.8765 |
| 346157 | ZNF391 | Zinc finger protein 282‐like (Peromyscus maniculatus bairdii) | 0.0174 | 9.7798 |
Table 6.
Top 20 annotated genes downregulated in BPA‐exposed males compared to control males
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 125950 | RAVER1 | Peromyscus maniculatus bairdii ribonucleoprotein, PTB‐binding 1 (Raver1), transcript variant X3, mRNA | 0.0453 | −11.3089 |
| 58508 | KMT2C | Histone‐lysine N‐methyltransferase 2C isoform X2 (Mus musculus) | 0.0358 | −11.1299 |
| 1149 | CIDEA | Cell death activator CIDE‐A (Peromyscus maniculatus bairdii) | 0.0343 | −10.9595 |
| 146547 | PRSS36 | Peromyscus maniculatus bairdii LysM, putative peptidoglycan‐binding, domain containing 4 (Lysmd4), mRNA | 0.0269 | −9.9781 |
| 11346 | SYNPO | Synaptopodin isoform X3 (Peromyscus maniculatus bairdii) | 0.0271 | −9.6247 |
| 1307 | COL16A1 | Collagen α‐1(XVI) chain isoform X4 (Peromyscus maniculatus bairdii) | 7.14E‐7 | −9.5970 |
| 84893 | FBXO18 | F‐box DNA helicase 1 isoform X1 (Peromyscus maniculatus bairdii) | 0.0270 | −9.5249 |
| 1663 | DDX11 | Probable ATP‐dependent RNA helicase DDX11 (Mesocricetus auratus) | 0.0079 | −9.4999 |
| 6445 | SGCG | Gamma‐sarcoglycan (Peromyscus maniculatus bairdii) | 0.0096 | −9.4984 |
| 7384 | UQCRC1 | Cytochrome b‐c1 complex subunit 1, mitochondrial (Peromyscus maniculatus bairdii) | 0.0352 | −9.4551 |
| 284098 | PIGW | Phosphatidylinositol‐glycan biosynthesis class W protein (Peromyscus maniculatus bairdii) | 0.0163 | −9.4045 |
| 131873 | COL6A6 | Collagen α‐4(VI) chain‐like (Peromyscus maniculatus bairdii) | 0.0407 | −9.3181 |
| 57698 | SHTN1 | Shootin‐1 isoform X1 (Peromyscus maniculatus bairdii) | 0.0005 | −9.2885 |
| 8732 | RGNTT | mRNA‐capping enzyme isoform X4 (Peromyscus maniculatus bairdii) | 0.0149 | −9.2616 |
| 2043 | EPHA4 | Low‐quality protein: ephrin type‐A receptor 10 (Lipotes vexillifer) | 0.0458 | −9.2506 |
| 284403 | WDR62 | WD repeat‐containing protein 62 isoform X4 (Peromyscus maniculatus) | 0.0250 | −9.2319 |
| 284111 | SLC13A5 | Solute carrier family 13 member 5 isoform X1 (Peromyscus maniculatus) | 0.0252 | −9.2208 |
| 138881 | OR1L8 | Mesocricetus auratus wntless homolog (Drosophila) (Wls), transcript variant X1, mRNA | 0.0085 | −9.1619 |
| 10114 | HIPK3 | Homeodomain‐interacting protein kinase 3 isoform X4 (Peromyscus maniculatus bairdii) | 0.0043 | −9.0835 |
| 11201 | POLI | DNA polymerase iota isoform X3 (Peromyscus maniculatus bairdii) | 0.0035 | −9.0237 |
Table 7.
Top 20 annotated genes upregulated in BPA‐exposed males compared to control males
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 84239 | ATP13A4 | Probable cation‐transporting ATPase 13A4 isoform X1 (Peromyscus maniculatus bairdii) | 1.76E‐07 | Double check |
| 26576 | SRPK3 | SRSF protein kinase 3 isoform X2 (Peromyscus maniculatus bairdii) | 0.0170 | 14.5735 |
| 583 | BBS2 | Bardet‐Biedl syndrome 2 protein (Peromyscus maniculatus bairdii) | 0.0176 | 13.6189 |
| 93099 | DMKN | Peromyscus maniculatus bairdii dermokine (Dmkn), mRNA | 0.0192 | 13.1855 |
| 26053 | AUTS2 | Autism susceptibility gene 2 protein isoform X5 (Peromyscus maniculatus bairdii) | 0.0218 | 12.8210 |
| 11335 | CBX3 | Peromyscus maniculatus bairdii AHA1, activator of heat‐shock 90kDa protein ATPase homolog 2 (yeast) (Ahsa2), mRNA | 0.0402 | 12.7135 |
| 64837 | KLC2 | Kinesin light chain 2 (Peromyscus maniculatus bairdii) | 0.0407 | 12.5940 |
| 222235 | FBXL13 | F‐box/LRR‐repeat protein 13 isoform X2 (Cricetulus griseus) | 0.0033 | 12.5725 |
| 10524 | KAT5 | Histone acetyltransferase KAT5 isoform X1 (Orcinus orca) | 0.0016 | 12.4747 |
| 9919 | SEC16A | Protein transport protein Sec16A isoform X2 (Peromyscus maniculatus bairdii) | 0.0338 | 12.1622 |
| 79041 | TMEM38A | Trimeric intracellular cation channel type A (Mesocricetus auratus) | 0.0437 | 12.0578 |
| 79041 | RTEL1 | Regulator of telomere elongation helicase 1 isoform X5 (Peromyscus maniculatus bairdii) | 0.0156 | 11.9364 |
| 51750 | KDM2A | Lysine‐specific demethylase 2A isoform X1 (Peromyscus maniculatus bairdii) | 0.0112 | 11.4489 |
| 22992 | ELMOD3 | ELMO domain‐containing protein 3 (Peromyscus maniculatus bairdii) | 0.0087 | 11.3341 |
| 84173 | OTUD7B | OTU domain‐containing protein 7B (Peromyscus maniculatus bairdii) | 0.0004 | 11.2632 |
| 56957 | DIXDC1 | Dixin isoform X7 (Ceratotherium simum simum) | 0.0022 | 11.0358 |
| 85458 | PCYOX1 | Prenylcysteine oxidase 1 (Peromyscus maniculatus bairdii) | 0.0450 | 10.9514 |
| 51449 | MAPK15 | Mitogen‐activated protein kinase 15 isoform X3 (Peromyscus maniculatus bairdii) | 0.0486 | 10.9511 |
| 225689 | SLC6A13 | Sodium‐ and chloride‐dependent GABA transporter 2 (Peromyscus maniculatus bairdii) | 0.0437 | 10.8735 |
| 27148 | KDM1B | Lysine‐specific histone demethylase 1B (Peromyscus maniculatus bairdii) | 0.0133 | 10.7690 |
Table 8.
Top 20 annotated genes downregulated in EE‐exposed males compared to control males
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 2805 | GOT1 | Aspartate aminotransferase | 0.0207 | −14.0902 |
| 23017 | FAIM2 | Protein lifeguard 2 isoform X2 (Tupaia chinensis) | 6.19E‐11 | −13.2226 |
| 6585 | SLIT1 | Slit homolog 1 protein (Peromyscus maniculatus bairdii) | 8.74E‐05 | −12.7204 |
| 51747 | LUC7L3 | Luc7‐like protein 3 isoform X1 (Microtus ochrogaster) | 0.0141 | −12.6836 |
| 152926 | PPM1K | Protein phosphatase 1K, mitochondrial (Peromyscus maniculatus bairdii) | 1.92E‐18 | −12.5670 |
| 26278 | SACS | Sacsin isoform X1 (Peromyscus maniculatus bairdii) | 0.0248 | −11.9637 |
| 11237 | RNF24 | Ring finger protein 24 (Cricetulus griseus) | 0.0177 | −11.9202 |
| 3992 | FADS1 | Fatty acid desaturase 1 isoform X2 (Peromyscus maniculatus bairdii) | 0.0188 | −11.8140 |
| 10207 | PATJ | InaD‐like protein isoform X4 (Peromyscus maniculatus bairdii) | 0.0005 | −11.2626 |
| 6650 | CAPN15 | Calpain‐15 isoform X1 (Peromyscus maniculatus bairdii) | 0.0009 | −11.2105 |
| 22866 | CNKSR2 | Connector enhancer of kinase suppressor of ras 2 isoform X4 (Peromyscus maniculatus bairdii) | 0.0017 | −10.9313 |
| 6196 | RPS6KA2 | Ribosomal protein S6 kinase α‐2 isoform X2 (Peromyscus maniculatus bairdii) | 0.0171 | −10.808 |
| 51430 | SUCO | Low‐quality protein: SUN domain‐containing ossification factor, partial (Peromyscus maniculatus bairdii) | 0.0107 | −10.5324 |
| 10507 | SEMA4D | Semaphorin‐4D isoform X4 (Peromyscus maniculatus bairdii) | 0.0110 | −10.5169 |
| 23378 | RRP8 | Ribosomal RNA‐processing protein 8 (Peromyscus maniculatus bairdii) | 0.0389 | −10.4778 |
| 124944 | C17ORF49 | Chromatin complexes subunit BAP18 isoform X3 (Cricetulus griseus) | 0.0045 | −10.0029 |
| 5165 | PDK3 | Pyruvate dehydrogenase kinase, isozyme 3 isoform X2 (Peromyscus maniculatus bairdii) | 0.0034 | −9.8266 |
| 84286 | TMEM175 | Endosomal/lysomomal potassium channel TMEM175 (Peromyscus maniculatus bairdii) | 0.0270 | −9.4840 |
| 255967 | PAN3 | PAB‐dependent poly(A)‐specific ribonuclease subunit PAN3 isoform X8 (Mus musculus) | 0.0094 | −9.4430 |
| 126119 | JOSD2 | Josephin‐2 isoform X1 (Cricetulus griseus) | 3.58E‐05 | −9.3777 |
Table 9.
Top 20 annotated genes upregulated in EE‐exposed males compared to control males
| Entrez ID | Gene symbol | Gene name | FDR | Log2 fold change |
|---|---|---|---|---|
| 30000 | TNPO2 | Transportin‐2 (Mesocricetus auratus) | 0.0016 | 12.5441 |
| 6196 | RPS6KA21 | Ribosomal protein S6 kinase α‐2 isoform X2 (Peromyscus maniculatus bairdii) | 2.59E‐10 | 12.4266 |
| 2036 | EPB41L1 | Band 4.1‐like protein 1 isoform X12 (Peromyscus maniculatus bairdii) | 0.0022 | 12.2511 |
| 84918 | LRP11 | Low‐density lipoprotein receptor‐related protein 11 (Peromyscus maniculatus bairdii) | 0.0024 | 12.1854 |
| 386675 | KRTAP10‐7 | Cricetulus griseus protein kinase C, α (Prkca), transcript variant X1, mRNA | 0.0032 | 11.8746 |
| 84893 | FBXO18 | F‐box DNA helicase 1 isoform X1 (Peromyscus maniculatus bairdii) | 1.08E‐05 | 11.7283 |
| 9026 | HIP1R | Huntingtin‐interacting protein 1‐related protein isoform X1 (Mesocricetus auratus) | 0.0041 | 11.5967 |
| 79411 | GLB1L | β‐Galactosidase‐1‐like protein isoform X1 (Peromyscus maniculatus bairdii) | 2.05E‐5 | 11.3220 |
| 4043 | LRPAP1 | α‐2‐Macroglobulin receptor‐associated protein (Peromyscus maniculatus bairdii) | 2.42E‐5 | 11.2051 |
| 56927 | GPR108 | Protein GPR108 (Peromyscus maniculatus bairdii) | 0.0061 | 11.1658 |
| 22800 | RRAS2 | Ras‐related protein R‐Ras2 (Elephantulus edwardii) | 0.0001 | 11.1491 |
| 113178 | SCAMP4 | Secretory carrier membrane protein 4, isoform CRA_c | 0.0068 | 11.0335 |
| 11346 | SYNPO | Synaptopodin isoform X3 (Peromyscus maniculatus bairdii) | 4.62E‐5 | 10.9740 |
| 51290 | ERGIC2 | Endoplasmic reticulum‐Golgi intermediate compartment protein 2 isoform X3 (Microtus ochrogaster) | 0.0004 | 10.9722 |
| 22826 | DNAJC8 | DnaJ homolog subfamily C member 8 isoform X2 (Peromyscus maniculatus bairdii) | 0.0002 | 10.9581 |
| 396 | ARHGDIA | Rho GDP‐dissociation inhibitor 1 isoform X2 (Cricetulus griseus) | 0.0090 | 10.7457 |
| 8848 | TSC22D1 | Peromyscus maniculatus bairdii TSC22 domain family, member 1 (Tsc22d1), transcript variant X2, mRNA | 0.0001 | 10.7014 |
| 11011 | TLK2 | Serine/threonine‐protein kinase tousled‐like 2 isoform X1 (Peromyscus maniculatus bairdii) | 0.0094 | 10.6909 |
| 2887 | GRB10 | growth factor receptor‐bound protein 10 isoform X2 (Peromyscus maniculatus bairdii) | 1.25E‐5 | 10.6643 |
| N/A | N/A | Peromyscus maniculatus bairdii ArfGAP with SH3 domain, ankyrin repeat and PH | 0.0099 | 10.6337 |
Analysis of the gene expression differences to examine for gene ontology (GO) and pathways that were significantly different only revealed one term GO: 0007017: microtubule‐based processes, where there were 42 genes and FDR <0.05. Genes involved in this process were significantly enriched in the EE males compared to control males, and BPA males compared to control males. No other GO terms or pathways were significantly enriched in any of the other comparisons.
Sex differences in hypothalamic gene expression
Tables S1 and S2 list the top 20 genes that are down‐ and upregulated, respectively, in control males versus control females. Tables S3 and S4 include the top 20 genes that are down‐ and upregulated, respectively, in BPA males versus BPA females. Tables S5 and S6 include the top 20 genes that are down‐ and upregulated, respectively, in EE males versus EE females. Similar to above, sex differences in down‐ and upregulated genes varied based on the treatment. In the case of the top 20 genes that were downregulated in males, the only ones that overlapped include Celf4 in control and BPA groups (shaded in Tables S1 and S3), and Kcnip4 in control and EE groups (bold in Tables S1 and S5). However, comparison of the top 20 genes that are downregulated in males compared to females in BPA and EE revealed no overlapping transcripts (Tables S3 and S5). For the top 20 gene that are upregulated in males versus females, Tbkbp1 was included in the control and BPA groups (shaded in Tables S2 and S4). No genes were shared among the top 20 genes that are upregulated in males versus females for control and EE and BPA and EE groups (Tables S2, S4, and S6).
Discussion
The goals of the current study were to determine if developmental exposure through the maternal diet to BPA or EE induces global gene expression changes in the hypothalamus of juvenile California mice. Second, we sought to determine whether the BPA and EE transcriptomic alterations were dependent on offspring sex. Since BPA is considered a weak estrogen (Vandenberg et al. 2009), the notion at the outset was that within sex similar gene expression pattern changes would be seen in BPA‐ and EE‐exposed individuals. The final aim was to examine for sexually dimorphic gene expression patterns within the control, BPA, and EE groups to determine if early exposure to one of the EDCs altered naturally occurring sex differences in hypothalamic gene expression in juvenile animals.
In relation to the first goal, RNA‐seq analysis identified several genes that were differentially expressed in the hypothalamus of BPA‐ and EE‐exposed females and males relative to their respective controls. Predictably, the specific gene expression patterns induced by each treatment varied based on offspring sex. A recent study examined gene expression patterns in the hippocampus and hypothalamus of PND1 rats exposed in utero to varying doses of BPA and EE, and reported only minimal gene expression changes (Arambula et al. 2016). As detailed above, this study design, however, failed to consider the postnatal exposure, which may be even more critical in rodent hypothalamic and other brain region development. It is this time period that approximates the third trimester in humans (Rice and Barone 2000; Howdeshell 2002). Additionally, in the previous study, the average number of reads per sample, presumably raw (although not explicitly specified) was only 29.2 million for the hypothalamus, which is about half of the total number of reads that were generated in the current studies. This difference is likely because the earlier study included more samples per lane, although no indication was provided on how many samples per lane were sequenced. In the current study, only three samples were sequenced per lane to ensure sufficient coverage per sample. Of the genes listed in the article, there does not appear to be any overlap with the current studies. Although the authors indicate a full set of differentially expressed hypothalamic genes included in Table S2a and S2b, these could not be accessed from the journal website nor could any of the other supplemental material with the message on this site reading: “We are sorry, this page is not available.” Thus, no additional comparisons to this earlier study can be made until the files become publicly available.
Although select genes were up‐ and downregulated in both BPA and EE females and males, in general, for both sexes unique genes were altered in BPA and EE groups. The findings thus provide further evidence that BPA can induce effects by binding to other steroidogenic and nonsteroidogenic receptors besides ESRs. Another interpretation of these data is that as a weak estrogen, BPA may not fully recapitulate the same gene expression changes as EE.
However, select genes were shared among highly differentially expressed genes in BPA‐ and EE‐exposed females and males. These transcripts will be further considered. Kcnd3 was among the most highly downregulated genes in the hypothalamus of both BPA‐ and EE‐exposed females. This gene encodes for the voltage‐gated potassium 4.3 (Kv4.3) channel. In humans, mutation of KCND3 is associated with cerebellar ataxia, intellectual disability, epilepsy, attention deficit disorders, and other clinical signs (Smets et al. 2015). Similar to the current findings, estrogen decreases transcriptional expression of Kv4.3 in the myometrium (Song et al. 2001).
Tranducin (beta)‐like 2 (Tbl2) was among the most highly upregulated genes for BPA‐ and EE‐exposed females. This gene encodes a protein that localizes to the endoplasmic reticulum (ER). Under ER stress conditions, it interacts with PKR‐like ER‐resident kinase (PERK), modulates protein expression of activating transcription factor 4 (ATF4), and associates with 60S ribosomal subunit with the net effect being alterations of specific proteins (Tsukumo et al. 2015). Topoisomerase I‐binding protein (Topors) is also one of the genes that showed the greatest expression increase in BPA‐ and EE‐exposed females. During oxidative stress, TOPRS dissociates from the H2AX protein and exerts an important role in chromatin reorganization during DNA repair and apoptosis (Seong et al. 2012). Little is known about the role of neural Kif13a, which is also upregulated in these two treatment groups. Deletion of this kinesin family motor protein in mice results in anxiety‐like behaviors, likely due to reduced neuronal transport of 5HT(1A) receptor (Zhou et al. 2013). Phactr2 is also upregulated in these two groups. Phosphatase and actin regulators (PHACTRs) are abundant in the brain where they are reported to mediate activity of protein phosphatase 1 and actin‐binding protein. Additionally, Phactr2 expression is elevated after traumatic brain injury (Kim et al. 2012). Taken together, the genes that are abundant in BPA‐ and EE‐exposed females appear to be ones that in many cases are upregulated under varying stressful conditions and may help to partially alleviate further cellular damage and ensuing behavioral disturbances.
In males, no genes were common among the 20 most highly differentiated genes expressed in both BPA‐ and EE‐exposed males. However, looking outside of these 20 genes, 9 and 72 transcripts were down‐ and upregulated, respectively, in both of these groups (Fig. 2). These shared transcripts are involved in a variety of functions, as shown in Supporting Information File 2.
Comparison of all the gene expression changes in females and males for BPA and EE groups only revealed one GO term to be enriched, microtubule‐based processes. With their pleiotrophic roles, microtubules are essential for proper neuronal function. Microtubules must respond quickly to environmental changes to provide structural support for the Golgi apparatus, axon guidance, dendrites, neurite outgrowth, interkinetic nuclear migration, and separation of chromatids during mitosis to list a few of the functions ascribed to these structures in neurons (Prokop 2013; Breuss and Keays 2014; Liu and Dwyer 2014; Sainath and Gallo 2015). The microtubule cytoskeleton works in conjunction with microfilaments (actin) and intermediate filaments to facilitate intracellular transport. Previous studies suggest that BPA exposure can disrupt microtubule‐associated proteins in hypothalamic neurons (Yokosuka et al. 2008; Iwakura et al. 2010) and other cell types (Nakagomi et al. 2001; Lehmann and Metzler 2004; Can et al. 2005; George et al. 2008). Ethinyl estradiol can also disrupt microtubular function (Epe et al. 1989; Sato et al. 1992).
In relation to the final aim, several genes were identified in all groups to be expressed in a sexually dimorphic pattern. However, there was minimal overlap of genes between the three groups, indicating that each treatment leads to a unique signature pattern within each sex. Alternatively, early exposure to BPA and EE altered the normal pattern of sex‐specific neurobehavioral programming. Other EDCs lead to sex‐dependent changes in the hypothalamus of largemouth bass (Micropterus salmoides) (Martyniuk et al. 2010, 2013) and rats (Walker et al. 2014).
While it would have been optimal to confirm the gene expression differences with qPCR, all of the RNA from the micropunch samples was used for the RNA‐seq analysis. Such studies will be performed in follow‐up studies, along with examining for protein expression differences. We also wish to examine other brain regions and non‐neural tissues to determine if there are common transcripts altered by developmental exposure to BPA and EE. Furthermore, it would be of interest to examine for phenotypic changes that are consistent with BPA‐induced microtubule dysfunction.
In conclusion, the current results show that perinatal exposure to BPA or EE mediates sex‐dependent changes in the hypothalamus of juvenile California mice. Furthermore, BPA and EE exposure results in unique signature of hypothalamic transcripts in males and females. The one common target of BPA and EE exposure may be microtubule‐based processes, which could lead to a wide range of pathophysiological effects in neuronal cells. The studies also demonstrate that even in juvenile animals there are normal sexually dimorphic differences in gene expression in the hypothalamus, but these may be disrupted by early exposure to either EDC tested. It remains to be determined whether the gene expression changes, including microtubule‐associated genes, are responsible for behavioral deficits observed in California mice and possibly by translation to humans.
Conflict of Interest
The authors declare no competing financial interests.
Supporting information
Table S1. Top 20 annotated genes downregulated in control males compared to control females. Shaded row is included in the BPA group (Table S3), whereas bold row is included in the EE group (Table S5).
Table S2. Top 20 annotated genes upregulated in control males compared to control females. Shaded row is included in the BPA group (Table 4).
Table S3. Top 20 annotated genes downregulated in BPA males compared to BPA females. Shaded row is included in the Control group (Table S1).
Table S4. Top 20 annotated genes upregulated in BPA males compared to BPA females. Shaded row is listed in controls (Table S2).
Table S5. Top 20 annotated genes downregulated in EE males compared to EE females. Bold row is included in the control group (Table S1).
Table S6. Top 20 annotated genes upregulated in EE males compared to EE females.
Data S1: Genes Increased or Decreased in Both BPA‐ and EE‐Exposed Groups.
Data S2: Differentially Expressed Genes Based on Treatment and Sex.
Data S3: Differentially Expressed Genes Based on Treatment.
Acknowledgments
The authors are grateful to Michele S. Painter and Angela B. Javurek for their assistance with the California mice colony, Leif A. McAllister for assistance in preparing the tables, Donald L. Connor for assistance with the figure preparation, and the DNA Core staff for assistance with the RNA validation methods and RNA‐seq procedure.
Johnson S. A., Spollen W. G., Manshack L. K., Bivens N. J., Givan S. A., Rosenfeld C. S.. Hypothalamic transcriptomic alterations in male and female California mice (Peromyscus californicus) developmentally exposed to bisphenol A or ethinyl estradiol, Physiol Rep, 5 (3), 2017, e13133, doi: 10.14814/phy2.13133
Funding Information
This project was funded by NIH Grant 5R21ES023150, the Mizzou Advantage Program, the Bond Life Sciences Center at the University of Missouri, and the University of Missouri, Office of Research.
Contributor Information
Scott A. Givan, Email: givans@missouri.edu
Cheryl S. Rosenfeld, Email: rosenfeldc@missouri.edu
References
- Lukasz Komsta 2011. Outliers: Tests for outliers. Available at: https://cran.r-project.org/web/packages/outliers/index.html KLoTfoIAf (accessed 19 January 2017).
- Adkins‐Regan, E. 2005. Hormones and animal social behavior. Princeton University Press, Princeton, NJ. [Google Scholar]
- Adler, D . MDrDVUOI. 2016. Available at: https://cran.r-project.org/web/packages/rgl/index.html. 2016. (accessed 19 January 2017)
- Altschul, S. F. , Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. 1997. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arambula, S. E. , Belcher S. M., Planchart A., Turner S. D., and Patisaul H. B.. 2016. Impact of low dose oral exposure to bisphenol A (BPA) on the neonatal rat hypothalamic and hippocampal transcriptome: A CLARITY‐BPA consortium study. Endocrinology 157:3856–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, A. P. , and Breedlove S. M.. 1985. Organizational and activational effects of sex steroids on brain and behavior: a reanalysis. Horm. Behav. 19:469–498. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, B. , Henare K., Thorstensen E. B., Ponnampalam A. P., and Mitchell M. D.. 2010. Transfer of bisphenol A across the human placenta. Am. J. Obstet. Gynecol. 202:393e391–393e397. [DOI] [PubMed] [Google Scholar]
- Berenbaum, S. , and Hines M.. 1992. Early androgens are related to childhood sex‐typed toy preferences. Psychol. Sci. 3:203–206. [Google Scholar]
- Biedermann, S. , Tschudin P., and Grob K.. 2010. Transfer of bisphenol A from thermal printer paper to the skin. Anal. Bioanal. Chem. 398:571–576. [DOI] [PubMed] [Google Scholar]
- Braun, J. M. 2012. Endocrine disrupting compounds, gonadal hormones, and autism. Dev. Med. Child Neurol. 54:1068. [DOI] [PubMed] [Google Scholar]
- Braun, J. M. , Kalkbrenner A. E., Just A. C., Yolton K., Calafat A. M., Sjodin A., et al. 2014. Gestational exposure to endocrine‐disrupting chemicals and reciprocal social, repetitive, and stereotypic behaviors in 4‐ and 5‐year‐old children: the HOME study. Environ. Health Perspect. 122:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuss, M. , and Keays D. A.. 2014. Microtubules and neurodevelopmental disease: the movers and the makers. Adv. Exp. Med. Biol. 800:75–96. [DOI] [PubMed] [Google Scholar]
- Calafat, A. M. , Ye X., Wong L. Y., Reidy J. A., and Needham L. L.. 2008. Exposure of the U.S. population to bisphenol A and 4‐tertiary‐octylphenol: 2003‐2004. Environ. Health Perspect. 116:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can, A. , Semiz O., and Cinar O.. 2005. Bisphenol‐A induces cell cycle delay and alters centrosome and spindle microtubular organization in oocytes during meiosis. Mol. Hum. Reprod. 11:389–396. [DOI] [PubMed] [Google Scholar]
- Cano‐Nicolau, J. , Vaillant C., Pellegrini E., Charlier T. D., Kah O., and Coumailleau P.. 2016. Estrogenic Effects of Several BPA Analogs in the Developing Zebrafish Brain. Front. Neurosci. 10:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. , Mickens J. A., McCaffrey K. A., Leyrer S. M., and Patisaul H. B.. 2012. Neonatal Bisphenol A exposure alters sexually dimorphic gene expression in the postnatal rat hypothalamus. Neurotoxicology 33:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. , Rebuli M. E., Rogers J., Todd K. L., Leyrer S. M., Ferguson S. A., et al. 2013. Prenatal bisphenol A exposure alters sex‐specific estrogen receptor expression in the neonatal rat hypothalamus and amygdala. Toxicol. Sci. 133:157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. , Joyner L., Mickens J. A., Leyrer S. M., and Patisaul H. B.. 2014. Sex‐specific Esr2 mRNA expression in the rat hypothalamus and amygdala is altered by neonatal bisphenol A exposure. Reproduction 147:537–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli, I. , Della Seta D., Fiorenzani P., Farabollini F., and Aloisi A. M.. 2007. Estrogenic chemicals at puberty change ERalpha in the hypothalamus of male and female rats. Neurotoxicol. Teratol. 29:108–115. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Zhou L., Bai Y., Zhou R., and Chen L.. 2014. Sex differences in the adult HPA axis and affective behaviors are altered by perinatal exposure to a low dose of bisphenol A. Brain Res. 1571:12–24. [DOI] [PubMed] [Google Scholar]
- Chen, Y. A. , Tripathi L. P., and Mizuguchi K.. 2016. An integrative data analysis platform for gene set analysis and knowledge discovery in a data warehouse framework. Database 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhangawala, S. , Rudy G., Mason C. E., and Rosenfeld J. A.. 2015. The impact of read length on quantification of differentially expressed genes and splice junction detection. Genome Biol. 16:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cock, M. , Maas Y. G., and van de Bor M.. 2012. Does perinatal exposure to endocrine disruptors induce autism spectrum and attention deficit hyperactivity disorders? Review Acta Paediatr. 101:811–818. [DOI] [PubMed] [Google Scholar]
- Consumer Reports 2012. Available at: http://news.consumerreports.org/baby/2012/07/fda-bans-bpa-from-baby-bottles-sippy-cups.html (accessed 19 January 2017)
- Diamanti‐Kandarakis, E. , Bourguignon J. P., Giudice L. C., Hauser R., Prins G. S., Soto A. M., et al. 2009. Endocrine‐disrupting chemicals: an Endocrine Society scientific statement. Endocr. Rev. 30:293–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietert, R. R. , Dietert J. M., and Dewitt J. C.. 2011. Environmental risk factors for autism. Emerg. Health Threats J. 4:7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environment Canada . 2008. Screening assessment for the challenge phenol, 4,4' ‐(1‐methylethylidene)bis‐ (bisphenol A) chemical abstracts service registry number 80‐05‐7. Pp. 1–107. Ministers of the Environment and of Health , Gatineau, QC, Canada. [Google Scholar]
- Epe, B. , Harttig U. H., Schiffmann D., and Metzler M.. 1989. Microtubular proteins as cellular targets for carcinogenic estrogens and other carcinogens. Prog. Clin. Biol. Res. 318:345–351. [PubMed] [Google Scholar]
- Eppsl‐I‐G‐N . 2015. [Internet] Eppsl‐I‐G‐N. Available at: http://www.ncbi.nlm.nih.gov/genome/?term=NC_001422.1. 2015. (accessed 23 January 2016)
- Fukushima, A. , Funabashi T., Kawaguchi M., Mitsushima D., and Kimura F.. 2007. Bisphenol A induces transforming growth factor‐beta3 mRNA in the preoptic area: a cDNA expression array and Northern blot study. Neurosci. Lett. 411:81–85. [DOI] [PubMed] [Google Scholar]
- Funabashi, T. , Kawaguchi M., and Kimura F.. 2001. The endocrine disrupters butyl benzyl phthalate and bisphenol A increase the expression of progesterone receptor messenger ribonucleic acid in the preoptic area of adult ovariectomized rats. Neuroendocrinology 74:77–81. [DOI] [PubMed] [Google Scholar]
- Galloway, T. , Cipelli R., Guralnick J., Ferrucci L., Bandinelli S., Corsi A. M., et al. 2010. Daily bisphenol A excretion and associations with sex hormone concentrations: Results from the InCHIANTI adult population study. Environ. Health Perspect. 118:1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary, D. C . 2010. Male, female : the evolution of human sex differences. Chapter 10. Pp. 291–294. American Psychological Association, Washington, DC. p. xv, 555 p. [Google Scholar]
- George, O. , Bryant B. K., Chinnasamy R., Corona C., Arterburn J. B., and Shuster C. B.. 2008. Bisphenol A directly targets tubulin to disrupt spindle organization in embryonic and somatic cells. ACS Chem. Biol. 3:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GitHub . FtI. Available at: https://github.com/agordon/fastx_toolkit (accessed 19 January 2017)
- Givan, S. A. , Bottoms C. A., and Spollen W. G.. 2012. Computational analysis of RNA‐seq. Methods Mol. Biol. 883:201–219. [DOI] [PubMed] [Google Scholar]
- Gonzalez, E. , and Joly S.. 2013. Impact of RNA‐seq attributes on false positive rates in differential expression analysis of de novo assembled transcriptomes. BMC Res. Notes 6:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean, P. , and Landrigan P. J.. 2006. Developmental neurotoxicity of industrial chemicals. Lancet 368:2167–2178. [DOI] [PubMed] [Google Scholar]
- GrandViewResearch . 2014. Global bisphenol A (BPA) market by application (appliances, automotive, consumer, construction, electrical & electronics) expected to reach USD 20.03 billion by 2020. Avaliable at: http://www.digitaljournal.com/pr/2009287 (accessed July 24 2014).
- Gross, J , and Ligges U.. 2015. nortest: Tests for Normality. Available from: https://cran.rproject.org/web/packages/nortest/index.html (accessed 19 January 2017)
- Gu, Z. , Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- Haas, B. J. , Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. 2013. De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. , Miao M., Herrinton L. J., Wu C., Yuan W., Zhou Z., et al. 2009. Bisphenol A levels in blood and urine in a Chinese population and the personal factors affecting the levels. Environ. Res. 109:629–633. [DOI] [PubMed] [Google Scholar]
- Howdeshell, K. L. 2002. A model of the development of the brain as a construct of the thyroid system. Environ. Health Perspect. 110(Suppl 3):337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezuki, Y. , Tsutsumi O., Takai Y., Kamei Y., and Taketani Y.. 2002. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum. Reprod. 17:2839–2841. [DOI] [PubMed] [Google Scholar]
- Iwakura, T. , Iwafuchi M., Muraoka D., Yokosuka M., Shiga T., Watanabe C., et al. 2010. In vitro effects of bisphenol A on developing hypothalamic neurons. Toxicology 272:52–58. [DOI] [PubMed] [Google Scholar]
- Jasarevic, E. , Sieli P. T., Twellman E. E., Welsh T. H. Jr, Schachtman T. R., Roberts R. M., et al. 2011. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol A. Proc. Natl Acad. Sci. USA 108:11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic, E. , Geary D., and Rosenfeld C.. 2012. Sexually selected traits: A fundamental framework for studies on behavioral epigenetics. ILAR J. 53:253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasarevic, E. , Williams S. A., Vandas G. M., Ellersieck M. R., Liao C., Kannan K., et al. 2013. Sex and dose‐dependent effects of developmental exposure to bisphenol A on anxiety and spatial learning in deer mice (Peromyscus maniculatus bairdii) offspring. Horm. Behav. 63:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. A. , Javurek A. B., Painter M. S., Ellersieck M. R., Welsh T. H. Jr, Camacho L., et al. 2015a. Effects of developmental exposure to bisphenol A on spatial navigational learning and memory in rats: A CLARITY‐BPA study. Horm. Behav. 80:139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. A. , Javurek A. B., Painter M. S., Peritore M. P., Ellersieck M. R., Roberts R. M., et al. 2015b. Disruption of parenting behaviors in California Mice, a monogamous rodent species, by endocrine disrupting chemicals. PLoS ONE 10:e0126284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. A. , Painter M. S., Javurek A. B., Ellersieck M. R., Wiedmeyer C. E., Thyfault J. P., et al. 2015c. Sex‐dependent effects of developmental exposure to bisphenol A and ethinyl estradiol on metabolic parameters and voluntary physical activity. J. Develop. Origins Health Dis. 6:539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. , Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., and Walichiewicz J.. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Kalkbrenner, A. E. , Schmidt R. J., and Penlesky A. C.. 2014. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Curr. Probl. Pediatr. Adolesc. Health Care 44:277–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, K. , Chauhan V., Gu F., and Chauhan A.. 2014. Bisphenol A induces oxidative stress and mitochondrial dysfunction in lymphoblasts from children with autism and unaffected siblings. Free Radic. Biol. Med. 76:25–33. [DOI] [PubMed] [Google Scholar]
- Kawamoto, Y. , Matsuyama W., Wada M., Hishikawa J., Chan M. P., Nakayama A., et al. 2007. Development of a physiologically based pharmacokinetic model for bisphenol A in pregnant mice. Toxicol. Appl. Pharmacol. 224:182–191. [DOI] [PubMed] [Google Scholar]
- Kim, J. Y. , Choi S. Y., Moon Y., Kim H. J., Chin J. H., Kim H., et al. 2012. Different expression patterns of Phactr family members in normal and injured mouse brain. Neuroscience 221:37–46. [DOI] [PubMed] [Google Scholar]
- Kundakovic, M. , Gudsnuk K., Franks B., Madrid J., Miller R. L., Perera F. P., et al. 2013a. Sex‐specific epigenetic disruption and behavioral changes following low‐dose in utero bisphenol A exposure. Proc. Natl Acad. Sci. USA 110:9956–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic, M. , Gudsnuk K., Franks B., Madrid J., Miller R. L., Perera F. P., et al. 2013b. Sex‐specific epigenetic disruption and behavioral changes following low‐dose in utero bisphenol A exposure. Proc. Natl Acad. Sci. USA 110:9956–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrigan, P. J. 2010. What causes autism? Exploring the environmental contribution. Curr. Opin. Pediatr. 22:219–225. [DOI] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell C., Pop M., and Salzberg S. L.. 2009. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, L. , and Metzler M.. 2004. Bisphenol A and its methylated congeners inhibit growth and interfere with microtubules in human fibroblasts in vitro. Chem. Biol. Interact. 147:273–285. [DOI] [PubMed] [Google Scholar]
- Li, B. , and Dewey C. N.. 2011. RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , and Dwyer T.. 2014. Microtubule dynamics in axon guidance. Neurosci. Bull. 30:569–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adaptor sequences from high‐throughput sequencing reads. EMBnet J. 17:10. [Google Scholar]
- Martyniuk, C. J. , Feswick A., Spade D. J., Kroll K. J., Barber D. S., and Denslow N. D.. 2010. Effects of acute dieldrin exposure on neurotransmitters and global gene transcription in largemouth bass (Micropterus salmoides) hypothalamus. Neurotoxicology 31:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk, C. J. , Doperalski N. J., Kroll K. J., Barber D. S., and Denslow N. D.. 2013. Sexually dimorphic transcriptomic responses in the teleostean hypothalamus: a case study with the organochlorine pesticide dieldrin. Neurotoxicology 34:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, M. M. 2008. Estradiol and the developing brain. Physiol. Rev. 88:91–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miodovnik, A. , Engel S. M., Zhu C., Ye X., Soorya L. V., Silva M. J., et al. 2011. Endocrine disruptors and childhood social impairment. Neurotoxicology 32:261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje, L. , Varayoud J., Luque E. H., and Ramos J. G.. 2007. Neonatal exposure to bisphenol A modifies the abundance of estrogen receptor alpha transcripts with alternative 5'‐untranslated regions in the female rat preoptic area. J. Endocrinol. 194:201–212. [DOI] [PubMed] [Google Scholar]
- Mueller, S. C. , Temple V., Oh E., VanRyzin C., Williams A., Cornwell B., et al. 2008. Early androgen exposure modulates spatial cognition in congenital adrenal hyperplasia (CAH). Psychoneuroendocrinology 33:973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi, M. , Suzuki E., Usumi K., Saitoh Y., Yoshimura S., Nagao T., et al. 2001. Effects of endocrine disrupting chemicals on the microtubule network in Chinese hamster V79 cells in culture and in Sertoli cells in rats. Teratog. Carcinog. Mutagen. 21:453–462. [DOI] [PubMed] [Google Scholar]
- NCfBI . [Internet]. NCfBI. Available at: https://www.ncbi.nlm.nih.gov/ (accessed 19 January 2017)
- Nishikawa, M. , Iwano H., Yanagisawa R., Koike N., Inoue H., and Yokota H.. 2010. Placental transfer of conjugated bisphenol A and subsequent reactivation in the rat fetus. Environ. Health Perspect. 118:1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanza, P. , Morellini, F. , Parmigiani, S. , and vom Saal, F. S. . 1999. Prenatal exposure to endocrine disrupting chemicals: effects on behavioral development. Neurosci. Biobehav. Rev. 23:1011–1027. [DOI] [PubMed] [Google Scholar]
- Paxinos, G . 2013. P. 472 in The rat brain in stereotaxic coordinates, 7th ed Academic Press, Cambridge, MA. [Google Scholar]
- Paxinos, G. A. F. , Franklin, K. B. J. 2008. P. 256 The mouse brain in stereotaxic coordinates, compact 3rd ed The coronal plates and diagrams. Academic Press, Cambridge, MA. [Google Scholar]
- Prokop, A. 2013. The intricate relationship between microtubules and their associated motor proteins during axon growth and maintenance. Neural. Develop. 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts, D. A. , McDaniel M. A., Jordan C. L., and Breedlove S. M.. 2008. Spatial ability and prenatal androgens: meta‐analyses of congenital adrenal hyperplasia and digit ratio (2D:4D) studies. Arch. Sex. Behav. 37:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, D. , and Barone S. Jr. 2000. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ. Health Perspect. 108(Suppl 3):511–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, C. A. , Martyniuk C. J., Annis M. L., Brumbaugh W. G., Chasar L. C., Denslow N. D., et al. 2014. Methylmercury‐induced changes in gene transcription associated with neuroendocrine disruption in largemouth bass (Micropterus salmoides). Gen. Comp. Endocrinol. 203:215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D. , McCarthy D. J., and Smyth G. K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld, C. S. 2015. Bisphenol A and phthalate endocrine disruption of parental and social behaviors. Front. Neurosci. 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vom Saal, F. S. , Richter C. A., Ruhlen R. R., Nagel S. C., Timms B. G., and Welshons W. V.. 2005. The importance of appropriate controls, animal feed, and animal models in interpreting results from low‐dose studies of bisphenol A. Birth Defects Res. A, Clin. Mol. Terat. 73: 140–145. [DOI] [PubMed] [Google Scholar]
- vom Saal, F. S. , Akingbemi B. T., Belcher S. M., Birnbaum L. S., Crain D. A., Eriksen M., et al. 2007. Chapel Hill bisphenol A expert panel consensus statement: integration of mechanisms, effects in animals and potential to impact human health at current levels of exposure. Reprod. Toxicol. 24: 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainath, R. , and Gallo G.. 2015. Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res. 359:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y. , Sakakibara Y., Oda T., Aizu‐Yokota E., and Ichinoseki K.. 1992. Effect of estradiol and ethynylestradiol on microtubule distribution in Chinese hamster V79 cells. Chem. Pharm. Bull. (Tokyo) 40:182–184. [DOI] [PubMed] [Google Scholar]
- Seong, K. M. , Nam S. Y., Kim J. Y., Yang K. H., An S., Jin Y. W., et al. 2012. TOPORS modulates H2AX discriminating genotoxic stresses. J. Biochem. Mol. Toxicol. 26:429–438. [DOI] [PubMed] [Google Scholar]
- Sieli, P. T. , Jasarevic E., Warzak D. A., Mao J., Ellersieck M. R., Liao C., et al. 2011. Comparison of serum bisphenol A concentrations in mice exposed to bisphenol A through the diet versus oral bolus exposure. Environ. Health Perspect. 119:1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smets, K. , Duarri A., Deconinck T., Ceulemans B., van de Warrenburg B. P., Zuchner S., et al. 2015. First de novo KCND3 mutation causes severe Kv4.3 channel dysfunction leading to early onset cerebellar ataxia, intellectual disability, oral apraxia and epilepsy. BMC Med. Genet. 16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. , Helguera G., Eghbali M., Zhu N., Zarei M. M., Olcese R., et al. 2001. Remodeling of Kv4.3 potassium channel gene expression under the control of sex hormones. J. Biol. Chem. 276:31883–31890. [DOI] [PubMed] [Google Scholar]
- Springer‐Verlag ; WHgEGfDAI. 2009. Available at: http://isbndb.com/book/ggplot2_elegant_graphics_for_data_analysis (accessed 23 January 2016).
- Stein, T. P. , Schluter M. D., Steer R. A., Guo L., and Ming X.. 2015. Bisphenol A exposure in children with autism spectrum disorders. Autism Res. 8:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukumo, Y. , Tsukahara S., Furuno A., Iemura S., Natsume T., and Tomida A.. 2015. The endoplasmic reticulum‐localized protein TBL2 interacts with the 60S ribosomal subunit. Biochem. Biophys. Res. Commun. 462:383–388. [DOI] [PubMed] [Google Scholar]
- Vandenberg, L. N. , Hauser R., Marcus M., Olea N., and Welshons W. V.. 2007. Human exposure to bisphenol A (BPA). Reprod. Toxicol. 24:139–177. [DOI] [PubMed] [Google Scholar]
- Vandenberg, L. N. , Maffini M. V., Sonnenschein C., Rubin B. S., and Soto A. M.. 2009. Bisphenol‐A and the great divide: a review of controversies in the field of endocrine disruption. Endocr. Rev. 30:75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg, L. N. , Chahoud I., Heindel J. J., Padmanabhan V., Paumgartten F. J., and Schoenfelder G.. 2010. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 118:1055–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D. M. , Goetz B. M., and Gore A. C.. 2014. Dynamic postnatal developmental and sex‐specific neuroendocrine effects of prenatal polychlorinated biphenyls in rats. Mol. Endocrinol. 28:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warita, K. , Mitsuhashi T., Ohta K., Suzuki S., Hoshi N., Miki T., et al. 2013. Gene expression of epigenetic regulatory factors related to primary silencing mechanism is less susceptible to lower doses of bisphenol A in embryonic hypothalamic cells. J. Toxicol. Sci. 38:285–289. [DOI] [PubMed] [Google Scholar]
- Warita, K. , Mitsuhashi T., Hoshi N., Ohta K., Suzuki S., Takeuchi Y., et al. 2014. A unique pattern of bisphenol a effects on nerve growth factor gene expression in embryonic mouse hypothalamic cell line N‐44. Arh Hig Rada Toksikol 65:293–299. [DOI] [PubMed] [Google Scholar]
- Williams, S. A. , Jasarevic E., Vandas G. M., Warzak D. A., Geary D. C., Ellersieck M. R., et al. 2013. Effects of developmental bisphenol A exposure on reproductive‐related behaviors in California mice (Peromyscus californicus): A monogamous animal model. PLoS ONE 8:e55698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbisky, S. E. , Weber G. J., Sepulveda M. S., Xiao C., Cannon J. R., and Freeman J. L.. 2015. Developmental origins of neurotransmitter and transcriptome alterations in adult female zebrafish exposed to atrazine during embryogenesis. Toxicology 333:156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C. E. , Rabaglino M. B., Richards E., Denslow N., Zarate M. A., Chang E. I., et al. 2014. Transcriptomics of the fetal hypothalamic response to brachiocephalic occlusion and estradiol treatment. Physiol. Genomics 46:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka, M. , Ohtani‐Kaneko R., Yamashita K., Muraoka D., Kuroda Y., and Watanabe C.. 2008. Estrogen and environmental estrogenic chemicals exert developmental effects on rat hypothalamic neurons and glias. Toxicol. In Vitro 22:1–9. [DOI] [PubMed] [Google Scholar]
- Zhou, R. , Niwa S., Guillaud L., Tong Y., and Hirokawa N.. 2013. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep. 3:509–519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Top 20 annotated genes downregulated in control males compared to control females. Shaded row is included in the BPA group (Table S3), whereas bold row is included in the EE group (Table S5).
Table S2. Top 20 annotated genes upregulated in control males compared to control females. Shaded row is included in the BPA group (Table 4).
Table S3. Top 20 annotated genes downregulated in BPA males compared to BPA females. Shaded row is included in the Control group (Table S1).
Table S4. Top 20 annotated genes upregulated in BPA males compared to BPA females. Shaded row is listed in controls (Table S2).
Table S5. Top 20 annotated genes downregulated in EE males compared to EE females. Bold row is included in the control group (Table S1).
Table S6. Top 20 annotated genes upregulated in EE males compared to EE females.
Data S1: Genes Increased or Decreased in Both BPA‐ and EE‐Exposed Groups.
Data S2: Differentially Expressed Genes Based on Treatment and Sex.
Data S3: Differentially Expressed Genes Based on Treatment.


