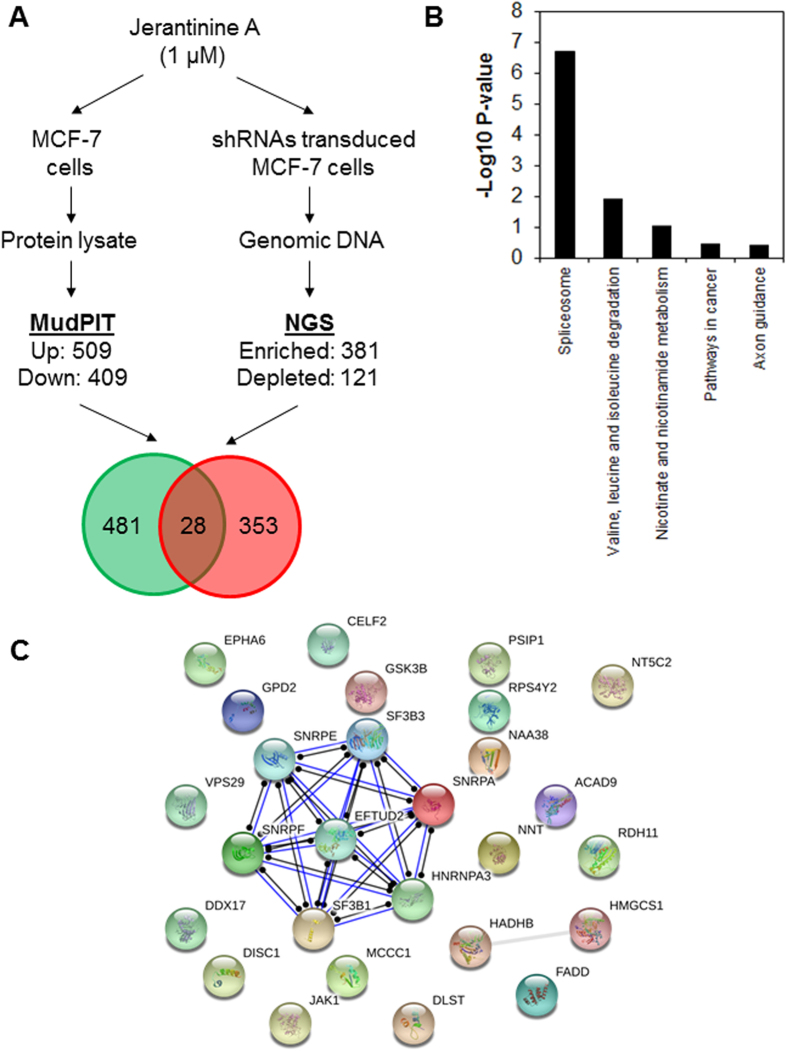
Figure 2. Combined proteomic-RNAi screen analyses revealed spliceosome as putative target of JA.
(A) Design of the combined proteomic profiling and genome-wide shRNA screen to identify new molecular targets of JA. MCF-7 cells were treated with 1 μM of JA for 8 h and their protein lysate were isolated for global proteomic analysis by MudPIT (left). Genes that were required for JA activities were identified by pooled genome-wide shRNA screen and NGS analysis following treatment of MCF-7 cells with 1 μM of JA for 72 h (right). The intersection of the data generated 28 candidate targets that were required for JA activities and were up-regulated in JA-treated cells. (B) DAVID enrichment analysis showing JA target genes associated with the spliceosome pathway (KEGG pathway, P < 0.01). (C) Protein-protein interaction network of the JA target proteins. Networks were generated with STRING at the highest confidence threshold (0.9). Note the seven interacting targets (SF3B1, SF3B3, SNRPA, SNRPE, SNRPF, HNRNPA3 and EFTUD2) are proteins within the spliceosome complex.