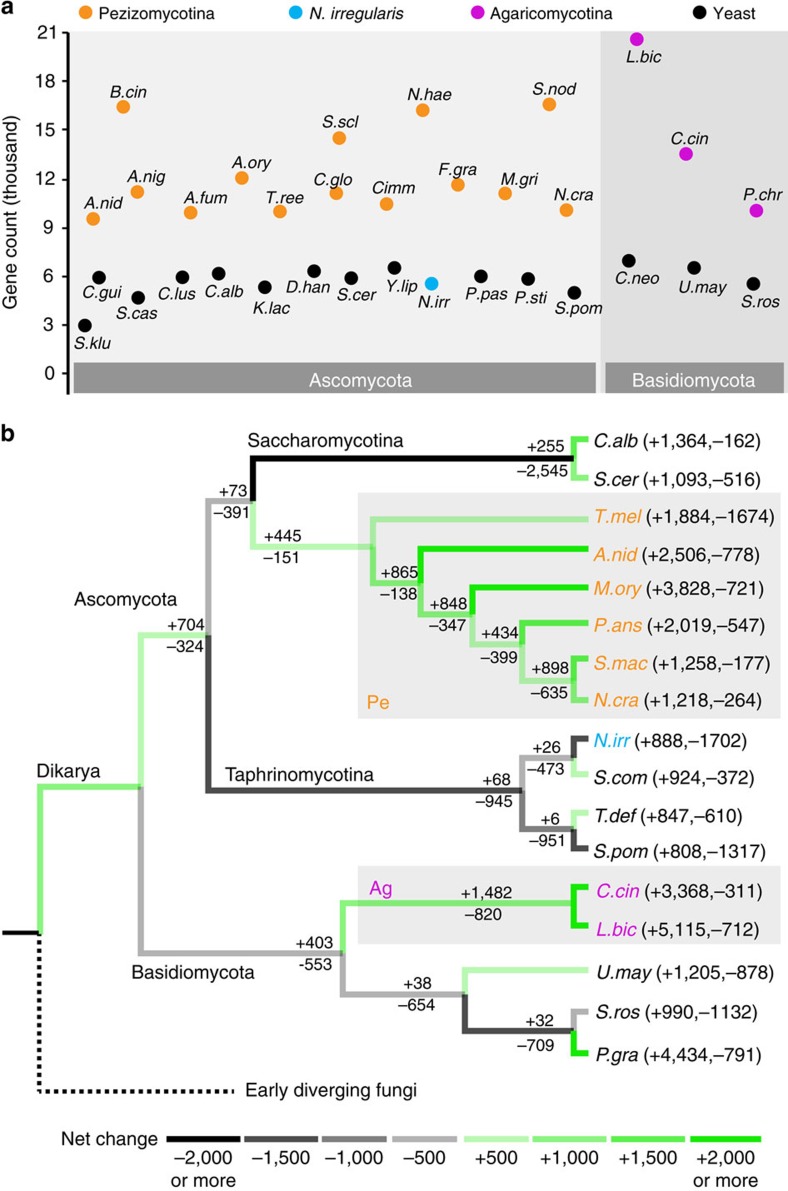
Figure 2. Estimated protein coding capacity of selected fungal genomes.
(a) The estimated number of genes is shown for selected members of the Ascomycota and Basidiomycota. A. fum, Aspergillus fumigatus. A. nig, Aspergillus niger. A. ory, Aspergillus oryzae. B. cin, Botrytis cinerea. C. gla, Candida glabrata. C. gui, Candida guilliermondii. C. lus, Candida lusitaniae. C. glo, Chaetomium globosum. C. imm, Coccidioides immitis. C. neo, Cryptococcus neoformans. D. han, Debaryomyces hansenii. F. gra, Fusarium graminearum. K. lac, Kluyveromyces lactis. M. gri, Magnaporthe grisea. N. hae, Nectria haematococca. P. pas, Pichia pastoris. P. sti, Pichia stipitis. S. cas, Saccharomyces castellii. S. klu, Saccharomyces kluyveri. S. scl, Sclerotinia sclerotiorum. S. nod, Stagonospora nodorum. T. ree, Trichoderma reesei. Y. lip, Yarrowia lipolytica. Other abbreviations are as indicated in the legend of Fig. 1. (b) Gains and losses of gene families at terminal taxa and internal nodes inferred using Dollo parsimony. Branch colour indicates the extent of net gain (green) or net loss (black) as defined in the legend.