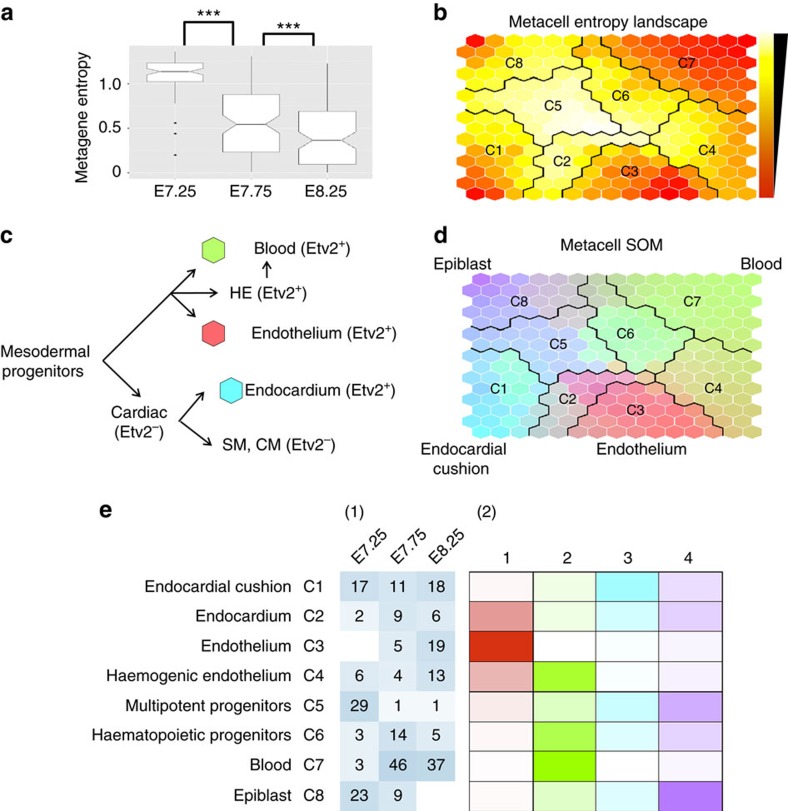
Figure 3. The metacell landscape and cluster analysis identified Etv2 derivatives.
(a) The cells from E7.25 had significantly higher metagene entropy than the cells from E7.75, and the metagene entropy of E7.75 cells was significantly higher than E8.25 cells (Wilcoxon rank-sum test, P value=1.2E-10 and P value=0.00075). (b) The distribution of metagene entropy of metacells is shown on the SOM. White colour represents high entropy metacells and red colour represents low entropy metacells. (c) A schematic represents a simplified version of the expected differentiation pathway with the dominant metagenes represented by colour for populations we expected to observe. (d) PAM algorithm clusters metacells by partitioning the metacell. The colour indicates the expression intensity of each metagene in the metacells. (e) Eight major cell clusters were identified by partitioning the metacell landscape. Each cell was mapped to the metacell with the most similar metagene coefficients. (1) The table indicates the time sources of cells from each cluster. (2) The heatmap shows the average metagene coefficients of each cell cluster.