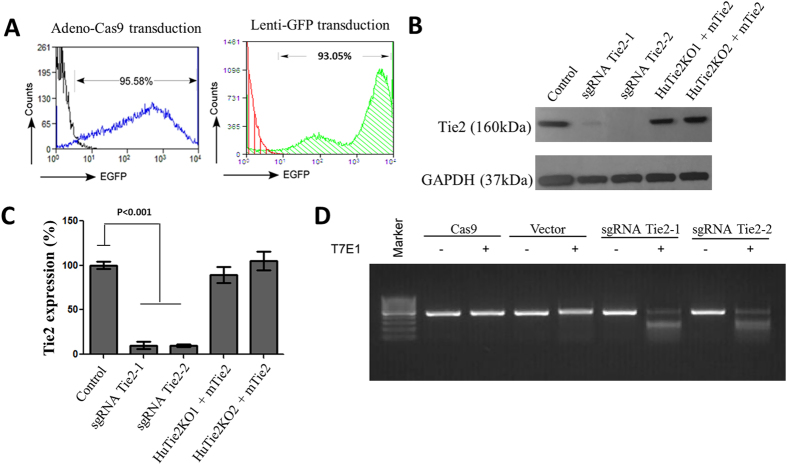
Figure 1. Deletion of Tie2 gene in primary ECs using CRISPR-Cas9.
(A) Cultured primary HLMVECs were transduced by EGFP-Cas9 adenovirus and sgRNA lentivirus targeting on Tie2 and subjected to flow cytometry analysis of EGFP expression. In a separate study, HLMVECs were transduced by GFP lentivirus and subjected to flow cytometry analysis of EGFP expression as an indicator of lentivirus transduction efficiency. (B) Protein expression of Tie2 in vector control and Tie2 knockout HLMVECs induced by CRISPR-Cas9 was determined by immunoblotting. Overexpression of mouse Tie2 in Tie2−/− HLMVECs was determined to assess the ability to rescue Tie2 expression after deletion. The uncropped full-length gels can be found in Supplementary Fig. S1. (C) Quantification of Tie2 protein expression from 3 independent experiments. sgRNA Tie2-1 and sgRNA Tie2-2 are CRISPR-Cas9-mediated deletions of Tie-2 at two distinction domains of Tie2 (two different sgRNAs). HuTie2KO1 and HuTie2KO2 + mTie2 represent restoration of Tie-2 expression in cells having undergone Tie-2 deletion. Differences were calculated using one-way ANOVA. P values less than 0.05 are indicated in the graph. (D) T7E1 assay detecting mutation on HLMVECs edited by CRISPR-Cas9 with Tie2 sgRNAs as showing a series of bands. Wild-types cells with or without Cas9 or vector only show single bands (negative controls).