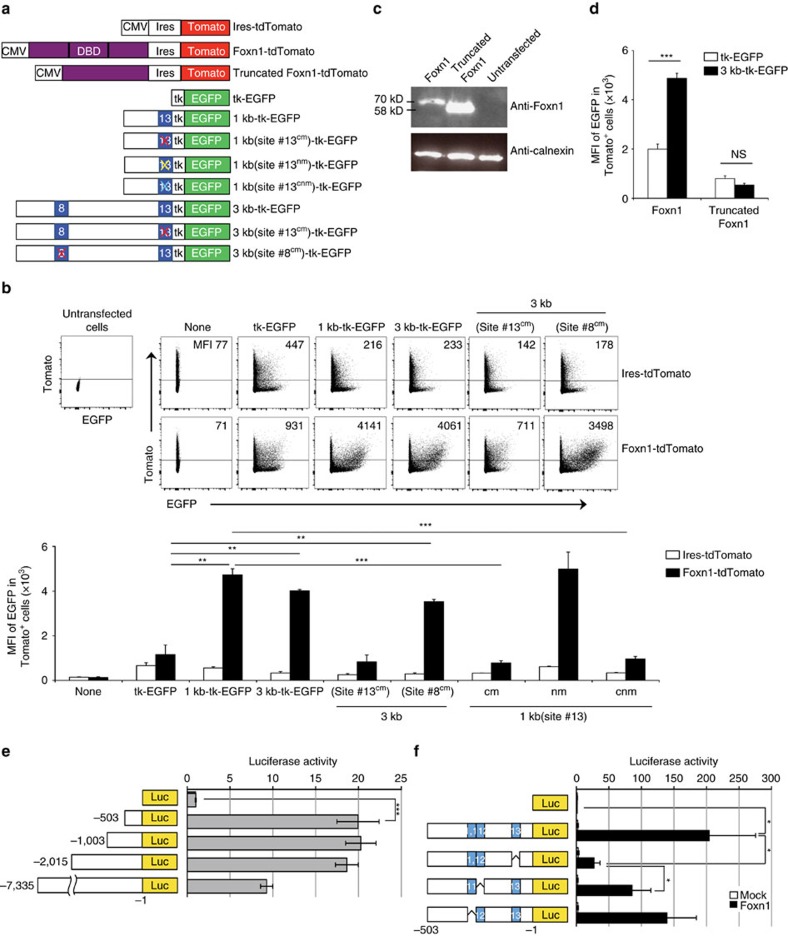
Figure 2. Foxn1 binding to site #13 enhances transcription of proximal gene.
(a) HEK293T cells were transfected with a plasmid that co-expressed Foxn1 protein and tdTomato red fluorescence protein and a plasmid that contained the EGFP green fluorescence protein reporter sequence attached to the herpes simplex virus thymidine kinase gene promoter (HSV-tk) and a variety of the mouse genomic sequence 5′ to β5t-encoding gene as indicated. Site #13 cm, a point mutation in the core sequence of site #13; site #13 nm, a point mutation in the non-core sequence of site #13; site #13 cnm, a mutation in both core and non-core sequence of site #13. (b) Dot plots show the expression of Tomato and EGFP in propidium iodide (PI)-negative viable HEK293T cells co-transfected with indicated EGFP reporter vectors and ires-tdTomato-expressing plasmid (upper profiles) or Foxn1-ires-tdTomato plasmid (lower profiles). Numbers in dot plots indicate mean fluorescence intensity (MFI) of EGFP expression in tdTomato+ PI- cells. Bar graphs show MFI (means±s.e.m., n=3) of EGFP expression in Tomato+ PI- cells. **P<0.01; ***P<0.001. See also Supplementary Fig. 2a. (c) Immunoblot analysis of Foxn1 protein or mutant Foxn1 protein without the DNA-binding domain (DBD). Calnexin was examined as the loading control. (d) HEK293T cells were co-transfected with indicated plasmids. EGFP reporter expression was measured as in b. ***P<0.005; n.s., not significant. (e) HEK293T cells were transfected with a series of luciferase reporter constructs that contained indicated lengths of the β5t 5′ genomic region, together with a Foxn1-encoding plasmid. Histograms represent relative luciferase activity, where the activity without genomic sequences was set as 1. Means±s.d. (n=3) are shown. ***P<0.005. (f) Luciferase reporter constructs that lacked site #11, #12 or #13 were tested. Means±s.d. (n=3) are shown. *P<0.05. All statistical analyses were performed by student's t-test.