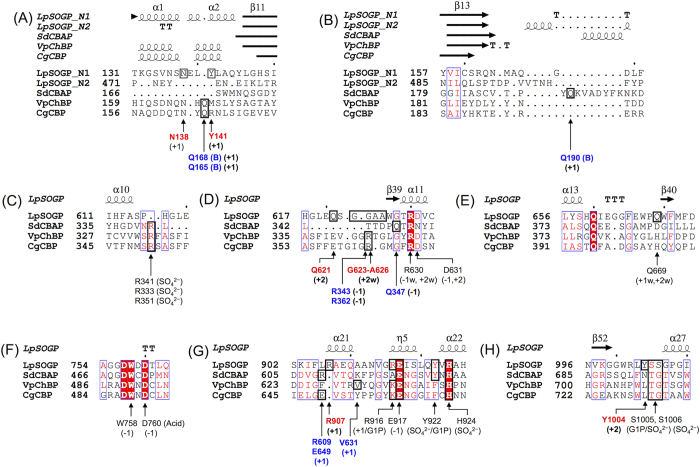
Figure 3. Multiple alignment of GH94 enzymes.
Multiple alignment was performed using Matras43. Figures were prepared using Espript 3.0 in the ENDscript server (http://espript.ibcp.fr)44. (A–B) Ndom1 and Ndom2 in LpSOGP, and Ndoms in SdCBAP, VpChBP, and CgCBP are aligned. (C–H) The catalytic domains of LpSOGP, SdCBP, VpChBP, and CgCBP are aligned. Residues related with substrate recognition are labeled, boxed, and indicated by arrows. Residues important for substrate binding at subsites +1 and +2 in LpSOGP are shown in red bold letters. Blue bold letters represent the residues in SdCBAP, VpChBP, and CgCBP that are located at the positions corresponding to the LpSOGP residues shown in red bold letters. Parentheses are the ligands with which the residues interact or the subsites where the residues bind to the ligands. Letters “w” in parentheses indicate that binding to the ligand is mediated by water molecules. (Acid) means that the residue is a catalytic acid. The residues derived from the B subunit are indicated by bold letter of B in parentheses.