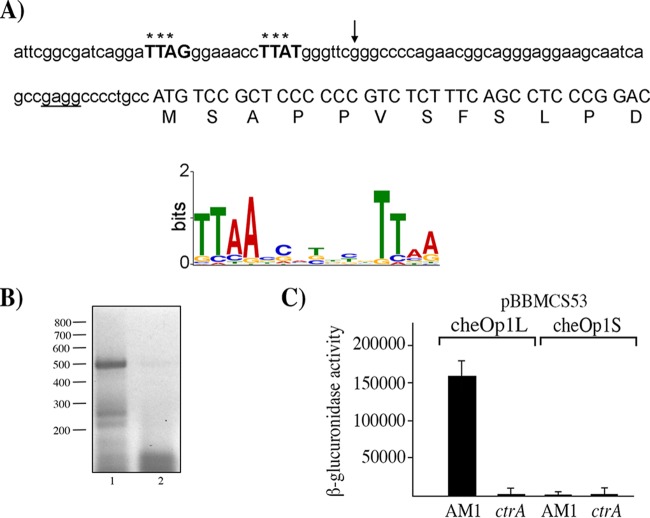
FIG 3.
Nucleotide sequence upstream of cheOp1, activity of the reporter gene uidA fused to different fragments from the regulatory region of cheOp1 in the pBBMCS53 plasmid, and RT-PCR data for cheY5 and its upstream gene. (A) Nucleotide sequence upstream of RSWS8N_02465. The first residues of the protein encoded by this gene are shown below the corresponding codons. The putative Shine-Dalgarno sequence is underlined, and nucleotides similar to the consensus binding site of CtrA, identified using a previously reported position weight matrix (31), are shown in bold and capital letters. The LOGO sequence of the CtrA-binding site is shown below; it was reported as part of a genomic analysis performed in C. crescentus (32). Asterisks above the sequence indicate nucleotides that match with the most frequent base in the LOGO sequence. An arrow indicates the 5′ end of the fragment cloned into pBBMCS53_cheOp1S. The 5′ end of the fragment cloned into pBBMCS53_cheOp1L includes 25 bp upstream of the sequence shown. (B) Agarose gel showing the RT-PCR products of the primer pair that amplifies the region encompassing CheY5 and RSWS8_02465. A 469-bp product was expected for this reaction. Lane 1, 5 μl from the RT-PCR; lane 2, 5 μl from the control reaction carried out in the absence of reverse transcriptase. The relevant molecular sizes from a 100-bp DNA ladder are shown on the left. (C) β-Glucuronidase activity of cell extracts from the AM1 and EA1 (ΔctrA::aadA) strains carrying the indicated plasmids. Cell cultures were grown in Sistrom's minimal medium supplemented with 80 μM succinic acid. Activity is reported in picomoles of 4-methylumbelliferone produced per minute per milligram of protein. The mean values and standard deviations (SD) of the results of three independent determinations are shown.