ABSTRACT
Baculovirus occlusion-derived virus (ODV) initiates infection of lepidopteran larval hosts by binding to the midgut epithelia, which is mediated by per os infectivity factors (PIFs). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) encodes seven PIF proteins, of which PIF1 to PIF4 form a core complex in ODV envelopes to which PIF0 and PIF6 loosely associate. Deletion of any pif gene results in ODV being unable to bind or enter midgut cells. AC83 also associates with the PIF complex, and this study further analyzed its role in oral infectivity to determine if it is a PIF protein. It had been proposed that AC83 possesses a chitin binding domain that enables transit through the peritrophic matrix; however, no chitin binding activity has ever been demonstrated. AC83 has been reported to be found only in the ODV envelopes, but in contrast, the Orgyia pseudotsugata MNPV AC83 homolog is associated with both ODV nucleocapsids and envelopes. In addition, unlike known pif genes, deletion of ac83 eliminates nucleocapsid formation. We propose a new model for AC83 function and show AC83 is associated with both ODV nucleocapsids and envelopes. We also further define the domain required for nucleocapsid assembly. The cysteine-rich region of AC83 is also shown not to be a chitin binding domain but a zinc finger domain required for the recruitment or assembly of the PIF complex to ODV envelopes. As such, AC83 has all the properties of a PIF protein and should be considered PIF8. In addition, pif7 (ac110) is reported as the 38th baculovirus core gene.
IMPORTANCE ODV is essential for the per os infectivity of the baculovirus AcMNPV. To initiate infection, ODV binds to microvilli of lepidopteran midgut cells, a process which requires a group of seven virion envelope proteins called PIFs. In this study, we reexamined the function of AC83, a protein that copurifies with the ODV PIFs, to determine its role in the oral infection process. A zinc finger domain was identified and a new model for AC83 function was proposed. In contrast to previous studies, AC83 was found to be physically located in both the envelope and nucleocapsid of ODV. By deletion analysis, the AC83 domain required for nucleocapsid assembly was more finely delineated. We show that AC83 is required for PIF complex formation and conclude that it is a true per os infectivity factor and should be called PIF8.
KEYWORDS: baculovirus, AcMNPV, AC83, PIF complex, per os infection, ZF domain, chitin binding domain, nucleocapsid assembly, occlusion-derived virus, fluorescence microscopy, PIF, zinc finger proteins
INTRODUCTION
Baculoviruses are double-stranded DNA enveloped viruses which specifically infect insects of the orders Lepidoptera, Hymenoptera, and Diptera (1). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) infects lepidopteran insects and is the most well-characterized baculovirus of the genus Alphabaculovirus. AcMNPV produces two virion phenotypes, occlusion-derived virus (ODV) and budded virus (BV), and each has a distinct role in the AcMNPV replication cycle. ODVs are required for per os infectivity and interhost transmission, whereas BVs cause systemic infection in an individual host (2). Although ODV and BV nucleocapsids have identical genetic contents, they are structurally and functionally distinct from each other. BVs have a single nucleocapsid enveloped in a membrane that is derived by budding from the host cell plasma membrane, whereas in ODVs, one or multiple nucleocapsids are enveloped in a membrane derived from the host cell nuclear envelope (3). ODVs become embedded in paracrystalline protein structures called occlusion bodies (OBs). OBs are liberated into the environment upon host liquefaction (2).
When ingested by larvae, OBs dissolve at the alkaline pH of the midgut, releasing ODV to infect the midgut epithelial cells (2). Upon release from OBs, ODVs must pass through the peritrophic matrix (PM) lining the host midgut in order to bind to receptors located on epithelial cells (4, 5). The binding of ODVs to midgut epithelial cells appears to have a high degree of specificity and requires a number of viral proteins, which includes the per os infectivity factors (PIFs) that are all known to be membrane proteins located in the ODV envelope. To date, seven PIF proteins have been identified, PIF0 (ac138; P74), PIF1 (ac119), PIF2 (ac22), PIF3 (ac115), PIF4 (ac96), PIF5 (ac148; ODV-E56), PIF6 (ac68), and PIF7 (ac110) (6–14). Until recently, all PIF proteins were encoded by core genes, which are genes conserved in all baculovirus genomes sequenced to date. An exception to this rule is the recently identified pif7, which has been reported to not be a core gene and is only found in the genomes of alpha- and betabaculoviruses (14). However, the overall high degree of conservation of these genes highlights the importance of PIF proteins in the evolution and biology of these viruses. Another common feature of all pif genes is that deletion results in loss of per os infectivity but does not impact BV production.
Recently, it was shown that PIF proteins form a high-molecular-weight PIF complex in the ODV envelope (15, 16). PIF1 to PIF4 associate to form the stable core of the PIF complex, and deletion of any of the genes encoding PIF1 to PIF4 results in the disruption of the PIF complex (16). Mass spectrometry and genetic data have also shown that PIF0 and PIF6, as well as AC5 and AC83, are associated with the PIF complex. PIF5 has been shown not to be associated with the core PIF complex (16). PIF0, PIF1, and PIF2 were reported to initiate binding of ODVs to epithelial cells (4, 10); however, functions of the other PIF proteins remain to be determined.
The PIF complex-associated protein, AC83 (P95), is 847 amino acids in length with a molecular mass of approximately 96 kDa and has significant similarities to PIF proteins. Like PIF proteins, AC83 is encoded by a core gene, contains a transmembrane domain, and copurifies with the PIF complex (13, 16). However, unlike pif genes, the deletion of ac83 results in the loss of nucleocapsid formation, and therefore no ODVs or BVs are produced (13, 17). Similar results were observed after deletion of the homologous gene bm95 in Bombyx mori NPV (BmNPV) (17, 18); however, deletion of specific AC83 domains can maintain BV production but oral infectivity is lost (13). AC83 was predicted to possess four structural domains, which include an inner nuclear membrane-sorting motif (INM-SM) at the N terminus containing a transmembrane (TM) domain, a C2H2 zinc finger (ZF) domain, a cysteine-rich type II chitin binding domain (CBD), and a short proline-rich (PR) domain (13, 17, 19). A region between the CBD and PR domains, which we have called the nucleocapsid assembly domain (NAD), has been shown to be required for nucleocapsid assembly (13). Deletion of the INM-SM, CBD, or PR domain did not impact BV production. Deletions of the AC83 CBD did, however, result in loss of ODV per os infectivity in Trichoplusia ni larvae (13). AC83 has, therefore, been proposed to disrupt the spatial structure of the peritrophic matrix to enable per os infection; however, in apparent contradiction to the proposed CBD function, chitin binding activity of AC83 has not been observed (13). Fractionation and proteomic analysis of AcMNPV and BmNPV ODV and BV has shown that AC83 localizes only to ODV, specifically the ODV envelope (13, 20, 21). However, in disagreement with these results, P91, which is the AC83 homolog in Orgyia pseudotsugata MNPV (OpMNPV), was detected in both ODV and BV. In addition, OpMNPV P91, in contrast to AcMNPV and BmNPV AC83, localizes to both nucleocapsids and envelope fractions of ODV when analyzed by immunoelectron microscopy and virion fractionation (22). OpMNPV P91 was also shown to have two nonglycosylated high-molecular-mass forms in ODV, but only the smaller one was present in BV. Due to the contrasting reports about the function and location of AC83 and its homologs, this study reexamined its role in the AcMNPV replication cycle. Using a series of AC83 deletion constructs, we show that AC83 is required for assembly or recruitment of the PIF complex to the ODV envelope. The AC83 assembly or recruitment function of the PIF complex requires the ZF domain, which is essential for oral infectivity. In addition, the NAD was further defined to a smaller region and is essential for BV production. Unlike previous studies, we show that full-length AC83 is located in both the ODV envelope and nucleocapsid, whereas the NAD only associates with the nucleocapsid. The core protein AC83, therefore, has the same properties as other PIF proteins with the additional function of being essential for nucleocapsid formation.
RESULTS
All pif genes are core genes.
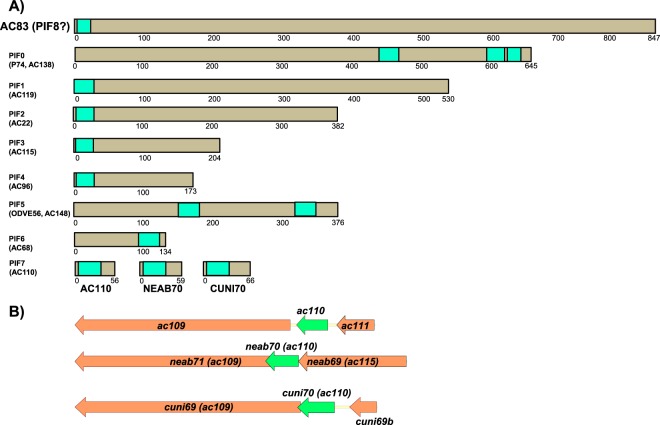
An earlier mass spectrometry study identified AC83 to be associated with the PIF complex (16), and it shares a number of common features with PIF proteins. All PIFs identified to date are encoded by core genes, contain one or more transmembrane domains (Fig. 1A), and are located in the ODV envelope. PIFs are essential for oral infectivity, and pif gene deletions do not impact BV production. The exception to these rules was the recently identified PIF7 (AC110), for which homologs were reported in all alpha- and betabaculoviruses, but it appeared to be absent from gamma- and deltabaculoviruses (14). Due to this apparent anomaly, we reexamined the available baculovirus genome data for ac110 homologs. This analysis revealed that all of the gamma- and deltabaculovirus genomes contain ac110 homologs located upstream of ac109 homologs, which is also a core gene (Fig. 1B). This makes ac110 a core gene like all other pif genes, and it is the 38th baculovirus core gene to be identified.
FIG 1.
Schematic diagrams showing per os infectivity (PIF) proteins. (A) A comparison of the relative sizes and locations of transmembrane domains of AC83 and the 8 known PIF proteins, PIF0 to PIF7. For AC110, the previously unreported homologs of AC110 found in the gamma- and deltabaculoviruses Neodiprion abeitis NPV (NEAB70) and Culex nigripalpus NPV (CUNI70) are also shown. (B) Schematic diagrams comparing the relative location of the pif7 ORF and its association with the core gene ac109 in AcMNPV, NeabNPV, and CuniNPV.
Reevaluation of the AC83 cysteine-rich region.
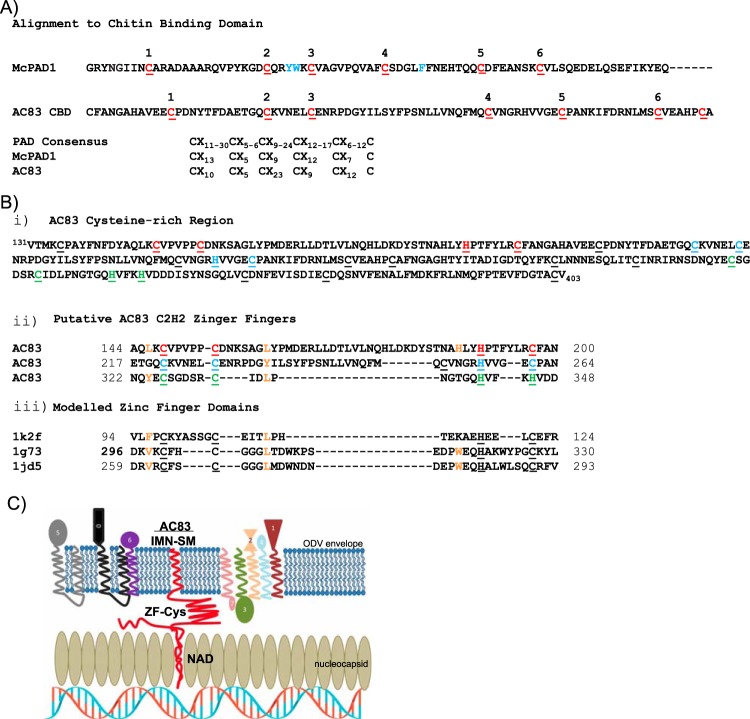
The AC83 cysteine-rich region, which is conserved in all ac83 orthologs, spans amino acids 131 to 403 and has previously been reported to contain a putative ZF between amino acids 148 to 197 and a CBD between amino acids 224 to 282 (Fig. 2A and B) (13). We reexamined this by comparing the AC83 CBD domain to known functional CBDs and other cysteine-rich domains. In agreement with earlier reports (13, 17), the region including amino acids 144 to 200 was predicted to possess a C2H2-type ZF (Fig. 2A and B). The position and separation of key cysteine and histidine residues involved in zinc coordination conformed to the consensus for these domains, and the region also contained residues adjacent to these that agreed with the looser zinc finger consensus of Krishna et al. (23). BLAST analysis of the region spanning amino acids 224 to 282 does return a domain report indicating that it contains a chitin binding peritrophin A domain (PAD; CBM_14 superfamily). This prediction, however, appears to be based only on an increased frequency of cysteine residues, a few of which conform to the PAD cysteine register consensus. We therefore performed homology modeling of this region to better ascertain its structure. A search of the protein structural database using SWISS-MODEL identified tachycitin, the archetype PAD, as a suitable template; however, only a short region (39 residues) could be modeled, and this had a low degree of statistical confidence (Q mean score of −1.61; sequence identity of 18.42%). Furthermore, alignment of this region to well-characterized chitin binding PADs revealed a lack of essential hydrophobic residues essential for chitin binding, namely, the hydrophobic residues between C2 and C3 in chitin binding module 1 and a hydrophobic residue between C4 and C5 in chitin binding module 2 (shown in blue in Fig. 2A). In addition, spacing of cysteine residues of AC83 does not conform to the six-cysteine register consensus of the highly conserved PAD sequences. These results indicate that the AC83 cysteine-rich domain is not a CBD. Closer inspection revealed that this region may in fact contain a second C2H2 ZF domain between amino acids 217 and 264 which was previously predicted for the AC83 ortholog BmP95 (19). In addition, a third ZF domain may reside between amino acids 322 and 348 (Fig. 2B). The AC83 cysteine-rich region from amino acids 114 to 348 would therefore appear to contain 3 tandem ZFs. A template search of the protein structural database with the individual putative ZF domains did not return any suitable templates; however, these are known to be difficult to model due to the high degree of diversity in the recognition region between individual ZFs (24).
FIG 2.
Analysis of the protein domains within the AC83 cysteine-rich region. (A) Alignment of the AC83 region predicted by Zhu et al. (13) to contain a type II peritrophin A chitin binding domain with a known representative PAD chitin binding domain from Mamestra configurata PAD1 (McPAD1; GenBank accession number GU596430) (51). Cysteine residues comprising the PAD domain six-cysteine register are shown in red. Essential hydrophobic residues involved in chitin binding are shown in blue. Comparison of the McPAD1 and AC83 cysteine registers to that of the expanded consensus for PAD domains according to Tetreau et al. (52) is shown below. (B, i) Amino acid sequence of the AC83 cysteine-rich region (amino acids 131 to 403) showing cysteine residues (underlined) and cysteine and histidine residues that may form ZF domains (red, blue, and green). (ii) Alignment of AC83 regions that may form C2H2-type ZF domains with (iii) representative ZF domains from proteins with known structures. Protein Data Bank codes are provided at the left. Zinc ligands are underlined. Additional residues conforming to the consensus of Krishna et al. (23) are shown in orange. (C) Proposed working model of AC83 function in ODV and interaction with PIF proteins. The model shows the schematic cross-section of an ODV nucleocapsid adjacent to the ODV envelope. The AC83 domains are shown associated with the nucleocapsid (NAD) and with the envelope (INM-SM), and the predicted zinc finger Cys-rich domain (ZF-Cys) is shown interacting with the core PIF complex (PIF1-4) and the associated PIF proteins (PIF0, PIF6, and PIF7) and PIF5.
AC83 deletion repair viruses are infectious.
Due to the previously published contradictory results about AC83 location and function, combined with our reanalysis of the cysteine-rich region, a new model was hypothesized for AC83 function (Fig. 2C). In this model, the inner nuclear membrane-sorting motif (INM-SM), which contains the transmembrane (TM) domain, anchors the AC83 protein in the ODV membrane. In addition, a domain required for nucleocapsid assembly (NAD) anchors AC83 in the nucleocapsid. The zinc finger domain, on the other hand, would be involved in recruiting or interacting with the PIF complex to the ODV membranes. One of the predictions of this model is that nucleocapsid assembly and PIF complex recruitment should be separable functions.
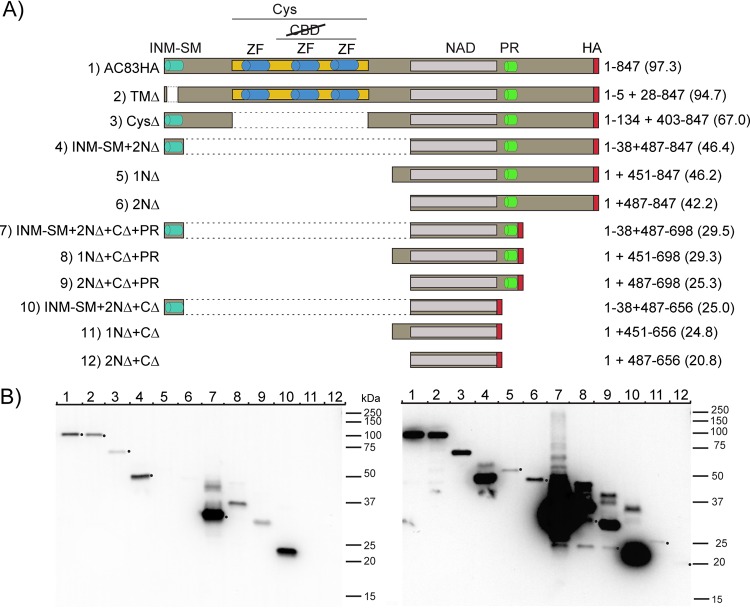
To test this hypothesis, a series of AC83 mutant viruses were constructed to examine impacts on AC83 expression, cellular localization, ODV virion location, BV production, in vivo per os infectivity, and interaction with the PIF complex. The series of AC83 mutants constructed are shown schematically in Fig. 3A. The mutants were constructed to examine the role and function of the AC83 domains, including the INM-SM and TM domains, the cysteine-rich ZF domain, the proline-rich domain (PR), and the N and C termini. In addition, we further defined the NAD that is required for wild-type (WT) levels of BV production (Fig. 3). All of the mutants were expressed in a bacmid backbone from which the wild-type AC83 had been deleted. Each construct was expressed under the control of the ac83 promoter and was tagged with the hemagglutinin (HA) epitope at the C terminus for detection of translated products by Western blotting.
FIG 3.
Construction and expression of AC83 deletions. (A) Schematic diagram showing AC83 domains and the proteins produced by each of the AC83 deletion constructs. Names of the viruses are shown on the left. The AC83 amino acids expressed from each construct are shown on the right with the predicted molecular mass, in kilodaltons, shown in parentheses. All proteins were tagged with the HA epitope at the C terminus. All constructs were expressed under the control of the ac83 promoter and inserted into an AcMNPV bacmid from which the wild-type AC83 had been deleted. Shown are the inner nuclear membrane sorting motif (INM-SM), cysteine-rich region (Cys), ZF C2H2 motif (ZF), chitin binding domain (CBD), nucleocapsid assembly domain (NAD), proline-rich domain (PR), and HA epitope tag (HA). (B) Western blot analysis of AC83 proteins expressed at 24 hpi in Sf9 cells infected at an MOI of 5. Proteins were detected using anti-HA antibody. Left and right panels represent short and long exposures, respectively, of the same blot, and each lane was loaded with an equal number of infected cells. The number in each lane corresponds to the number of the virus shown in panel A. Black dots have been placed beside the bands in either the left or right panel and correspond to predicted molecular mass.
All bacmid constructs expressing AC83 proteins were transfected into Spodoptera frugiperda clone 9 (Sf9) cells and were found to produce BV after bacmid transfection. Viral stocks were generated and used to infect cells to analyze the AC83 proteins produced (Fig. 3B). All of the AC83 peptides were expressed but at significantly varied levels. All of the constructs expressed a major protein at the expected molecular mass. In general, lower expression was observed with the constructs with both N- and C-terminal deletions (1NΔ, 2NΔ, 1NΔ+CΔ, and 2NΔ+CΔ). However, retention of the PR domain increased expression (compare 1NΔ+CΔ to 1NΔ+CΔ+PR or 2NΔ+CΔ to 2NΔ+CΔ+PR). The largest impact on expression was observed when the INM-SM was retained in the absence of the cysteine-rich region and the C terminus (INM-SM-2NΔ, INM-SM-2NΔ+CΔ+PR, and INM-SM-2NΔ+CΔ), as levels were observed that were many fold higher than that observed for the full-length protein (AC83-HA). In addition, the constructs that deleted the N terminus but retained the INM-SM or the PR domain produced multiple proteins of various molecular masses.
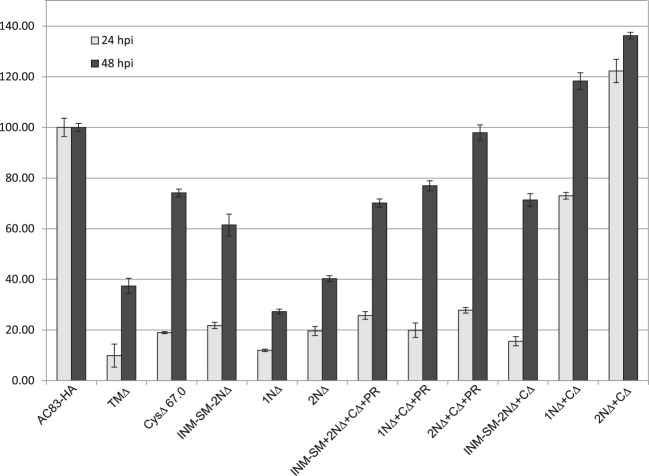
To assess the impact of the AC83 deletions on BV production, virus titers were determined at 24 and 48 h postinfection (hpi) for each construct (Fig. 4). At 24 hpi, all of the viruses produced BV levels lower than that of AC83-HA except for 2NΔ+CΔ, which produced 20% higher levels. The lowest levels at 24 hpi were observed for TMΔ, in which the TM domain was deleted from the INM-SM. At 48 hpi, 2NΔ+CΔ+PR, 1NΔ+CΔ, and 2NΔ+CΔ produced titers equal to or higher than that of AC83-HA. The remaining viruses all produced lower titers than that of AC83-HA. 2NΔ and 2NΔ+CΔ+PR produced lower BV titers than 2NΔ+CΔ, which suggests that the C terminus including the PR domain can inhibit BV production. These results clearly show that the majority of AC83 is not required for BV production and that the NAD, which is required for the production of wild-type or higher levels of BV, has been further defined to amino acids 487 to 656. Interestingly, 2NΔ+CΔ, which produced the highest BV titer, expressed the lowest AC83 levels (Fig. 3B).
FIG 4.
Relative BV production by AC83 deletion repair constructs in Sf9 cells at 24 and 48 hpi. Sf9 cells were infected with AC83HA, TMΔ, CysΔ, INM-SM+2NΔ, 1NΔ, 2NΔ, INM-SM+2NΔ+CΔ+PR, 1NΔ+CΔ+PR, 2NΔ+CΔ+PR, INM-SM+2NΔ+CΔ, 1NΔ+CΔ, and 2NΔ+CΔ (Fig. 3) at an MOI of 10. The BV titers in cell supernatants at 24 and 48 hpi were determined by droplet digital PCR.
Localization of AC83 and the AC83 NAD in ODV and BV.
Previous studies reported that AC83 or its homologs are located solely in the ODV envelope or alternatively in both the ODV envelope and the nucleocapsid. Based on our hypothesized model of AC83 function, the NAD that was mapped by the deletion constructs should be found only in the nucleocapsid fraction when expressed alone, whereas the full-length AC83 should be located in both the envelope and nucleocapsid. To test this, we prepared OBs from the AC83HA and 1NΔ+CΔ viruses, isolated the ODV, and separated the virions into envelope and nucleocapsid fractions (Fig. 5A). For the AC83-HA ODV, it was observed that the majority of AC83 was found in the envelope, but a small amount was also detected in the nucleocapsid fraction. The fractionation was repeated for 1NΔ+CΔ ODV, which only contains the mapped NAD, and unlike full-length AC83HA, protein was detected only in the nucleocapsid fraction and none in the envelope fraction. These results clearly show that AC83 is located in both the envelope and nucleocapsid of ODV with the region essential for BV development located in the nucleocapsid.
FIG 5.
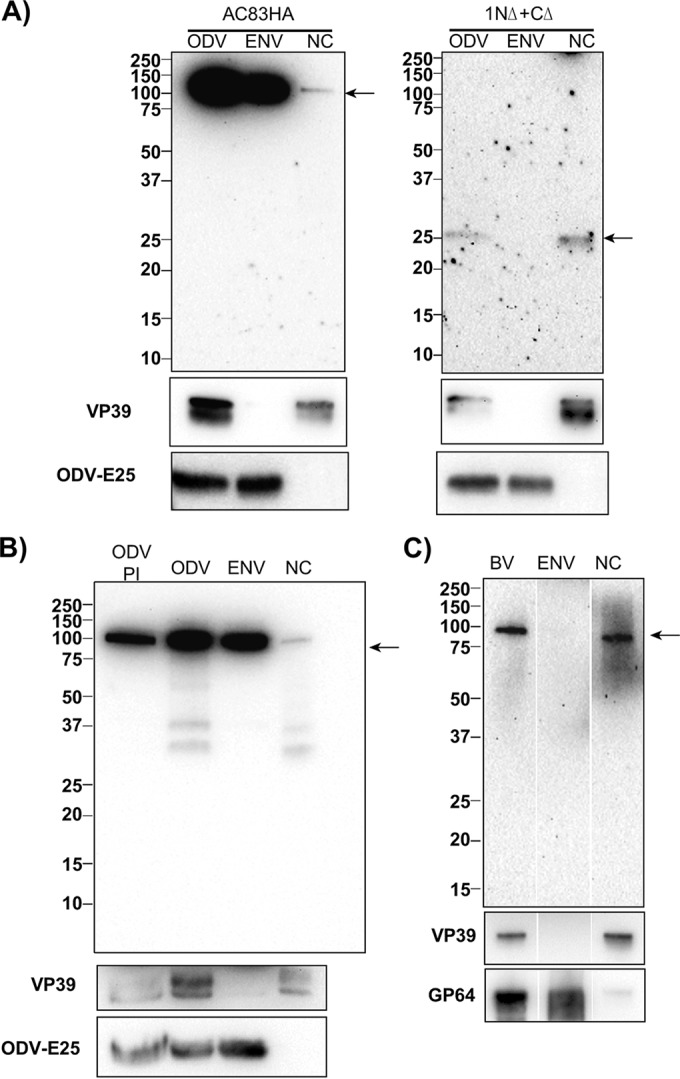
Localization of AC83HA and AC83 deletion 1NΔ+CΔ by Western blotting in ODV and BV envelope and nucleocapsid fractions. (A, left) Detection of full-length AC83 in total ODV (ODV) and envelope (ENV) and nucleocapsid (NC) fractions using anti-HA antibody. The majority of AC83HA is located in the ODV envelope, but a reduced amount is always found in the nucleocapsid fraction. (Right) Comparison of the fractionation of AC83HA to that of 1NΔ+CΔ. Unlike AC83HA, 1NΔ+CΔ AC83 is only located in the nucleocapsid fraction. (B) Western blot analysis of AC83HA isolated from ODV in the presence (PI) and absence of protease inhibitors. The blot shows that some of the AC83HA in the nucleocapsid fraction is cleaved in the absence of protease inhibitors. (C) Western blot analysis of BV as well as the envelope and nucleocapsid fractions of BV. The quality of the fractionation shown in panels A, B, and C was confirmed by Western blotting of the same samples using antisera against the known nucleocapsid protein VP39 and the ODV envelope protein ODV-E25 or the BV envelope protein GP64 (shown at the bottom).
Previously, it was shown that when OBs are isolated from insect larvae as opposed to tissue culture, PIF0 (P74) was cleaved into fragments when ODVs were released (25). We analyzed ODV isolated from tissue culture-derived OBs in the presence and absence of protease inhibitors. It was observed that specific AC83 cleavage products were detectable but only in the absence of protease inhibitors (Fig. 5B). Interestingly, fractionation of ODV in the absence of proteinase inhibitors resulted in only the nucleocapsid fraction producing the cleavage products.
Previous studies failed to identify AC83 in BV by Western blotting (AcMNPV and BmNPV [13, 17]) or proteomics analysis (AcMNPV and HearNPV [21, 26]); however, Russell and Rohrmann (22) showed by Western blot analysis that the AC83 homolog was located in OpMNPV BV. We therefore reanalyzed purified BV produced by AC83HA by Western blot analysis (Fig. 5C) and showed that with this virus, AC83 was copurified with BV. However, in contrast to ODV, the AC83 in BV fractionated almost entirely in the nucleocapsid.
Per os infectivity of AC83 mutants.
To examine the relationship of AC83 with the PIF complex, a subset of the ac83 mutants was analyzed for in vivo infectivity and interaction with the PIF complex. Our model predicts that the cysteine-rich ZF domain is responsible for interacting with the PIF complex. Therefore, we bioassayed viruses that expressed AC83 mutants with N-terminal changes covering the cysteine-rich zinc finger and the INM-SM domains, which included TMΔ, CysΔ, 2NΔ, and INM-SM-2NΔ. OBs of each of these viruses were generated in T. ni larvae. The presence of ODVs in OBs of all of these recombinant viruses was confirmed by Western blotting using an antibody recognizing the major AcMNPV capsid protein VP39 (Fig. 6). Western blot analysis of ODV purified from each virus detected approximately equal levels of VP39. This result confirmed that OBs of these recombinant viruses contained ODVs and therefore were used for in vivo bioassays.
FIG 6.
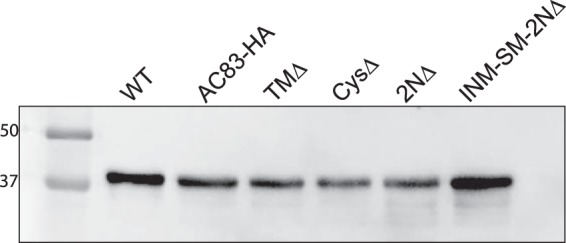
Analysis of ODV content in OBs isolated from T. ni larvae infected with WT, AC83HA, TMΔ, CysΔ, 2NΔ, and INM-SM+2NΔ. ODV were isolated from 1 × 107 OBs, and proteins were separated on a 10% SDS-PAGE gel and analyzed by Western blotting using an anti-VP39 antibody to detect the major nucleocapsid protein.
Oral infectivity of AC83HA, TMΔ, CysΔ, 2NΔ, and INM-SM-2NΔ, as well as WT virus, was tested in bioassays using T. ni 4th-instar larvae. WT and AC83HA viruses showed similar per os infectivity (90% lethal dose [LD90] of 44 and 31 OBs, respectively) (Table 1), which confirmed that oral infectivity of AC83HA was not compromised. Oral infectivity of TMΔ (LD90 of 480 OBs) was reduced approximately 10-fold compared to those of AC83HA and WT, which suggested that the transmembrane domain of AC83 plays a role in the efficiency of the oral infectivity of the virus; however, it is not absolutely required for per os infection. Oral infectivity for CysΔ, 2NΔ, and INM-SM-2NΔ was absolutely abolished even at very high OB doses (106 OBs/larva) (Table 1). The common feature of these three viruses was the absence of the AC83 cysteine-rich zinc finger domain of the protein, indicating that it is essential for oral infectivity of AcMNPV.
TABLE 1.
Percent mortality upon oral infection with OBs of 4th-instar T. ni larvae at 6 days pi
| Virus | Mortality (%) with oral dose (OBs) (n = 24) ofa: |
LD90 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 10 | 102 | 103 | 104 | 103 | 104 | 105 | 106 | ||
| WT | 42 | 100 | 100 | 100 | ND | ND | ND | ND | 44 |
| vAC83HA | 66 | 100 | 100 | 100 | ND | ND | ND | ND | 31 |
| vTMΔ | 12 | 21 | 100 | 100 | ND | ND | ND | ND | 480 |
| vCysΔ | ND | ND | ND | ND | 0 | 0 | 0 | 0 | |
| v2NΔ | ND | ND | ND | ND | 0 | 0 | 0 | 0 | |
| vINM-SM+2NΔ | ND | ND | ND | ND | 0 | 0 | 0 | 0 | |
ND, not done.
The bioassays with these N-terminal AC83 mutants showed that AC83 is required for oral infectivity. To determine if these AC83 mutants can cause systemic infection if the midgut is bypassed, BV was assayed by intrahemoceolic injection. BV from AC83HA, TMΔ, CysΔ, 2NΔ, INM-SM-2NΔ, and the WT were injected into 4th-instar T. ni larvae, and mortality was scored at 6 days postinjection (Table 2). All viruses exhibited 100% mortality, showing that the ability to cause a systemic infection is unaffected by deletion of the AC83 N terminus and loss of per os infectivity. This phenotype is the same as that previously observed with PIF0 to PIF7 gene deletions (6–14).
TABLE 2.
Percent mortality upon intrahemoceolic injection of BV in 4th-instar T. ni larvae
| Virus | Mortality (%) at 6 days pi (dose of 100 TCID50; n = 12) |
|---|---|
| vWT | 100 |
| vAC83HA | 100 |
| vTMΔ | 100 |
| vCysΔ | 100 |
| v2NΔ | 100 |
| vINM-SM+2NΔ | 100 |
| Grace's medium | 0 |
As described above, previous studies suggested that the AC83 cysteine-rich zinc finger domain was a CBD and was required for viral infection due to its interaction with either the PM or chitin-synthesizing cells of the midgut or the chitin-rich tracheal system. However, the model proposed in the current study suggests the cysteine-rich ZF domain is essential for PIF complex formation in ODV and would be critical for ODV binding and/or infection of the midgut epithelial cells. To investigate whether midgut infection is initiated upon oral ingestion of OBs, AC83HA was compared to CysΔ and INM-SM-2NΔ. T. ni 4th-instar larvae were orally infected with 104 OBs per larva and dissected to remove the midgut at 24, 48, and 72 hpi. Fluorescence microscopy of the isolated midgut tissue from these infected larvae showed green fluorescent protein (GFP) expression in AC83HA-infected larvae only; no GFP expression was detected in CysΔ- or INM-SM+2NΔ-treated larvae at 24, 48, or 72 hpi (Fig. 7). These data indicate that T. ni midgut infection is not initiated upon ingestion of CysΔ and INM-SM+2NΔ OBs. Thus, the role of AC83 is the establishment of oral infection in the larval host by enabling ODV to bind to or enter midgut cells.
FIG 7.
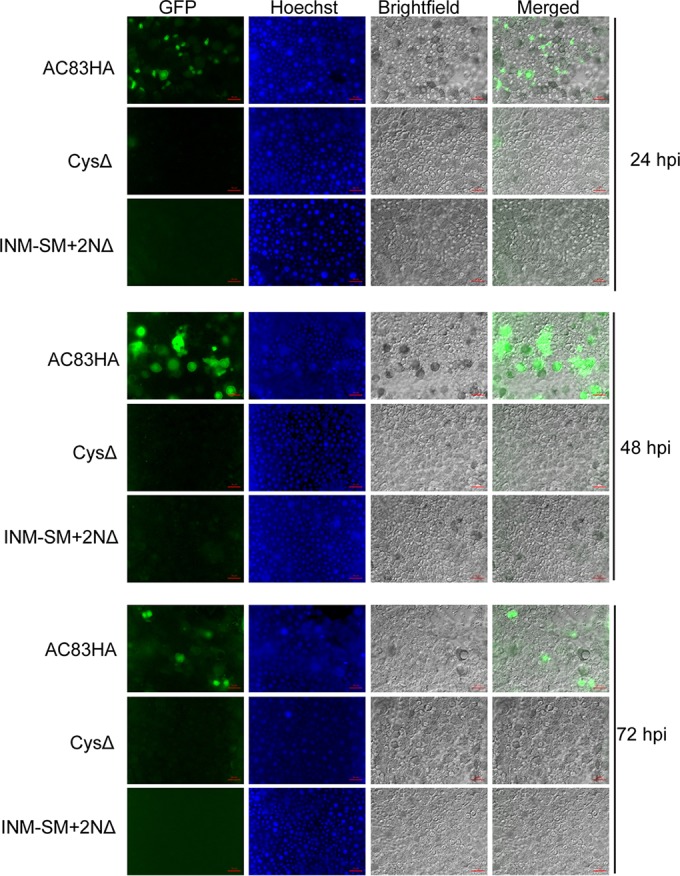
Infection of T. ni midgut by AC83HA, CysΔ, and INM-SM+2NΔ at 24, 48, and 72 hpi. T. ni larvae were orally fed with 104 OBs, and midgut epithelial cell layers were isolated at 24, 48, and 72 hpi. Midguts were observed by fluorescence microscopy for GFP expression as a marker for infection. Infection was observed in the midgut of larvae infected with AC83HA OBs but was not observed with CysΔ or vINM-SM+2NΔ. Midgut cell nuclei were stained with Hoechst 33342.
AcMNPV PIF complex formation is severely impaired in the absence of the ZF region of AC83.
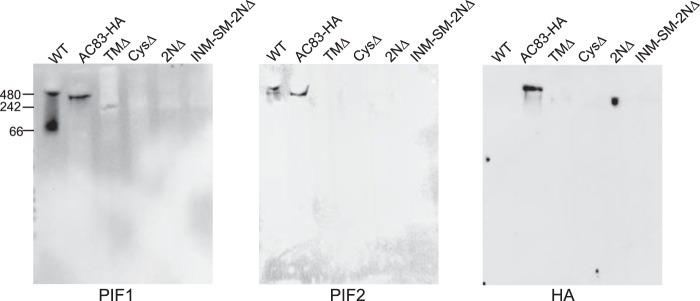
Previous studies showed that ODV envelopes contain the PIF complex, which includes PIF0 to PIF4 as well as AC83. Our data showed that AC83 was located in both the nucleocapsid and the envelope of ODV (Fig. 5). Our model predicts that AC83 via the cysteine-rich ZF domain is required for recruiting or assembly of the PIF complex to the ODV envelope, and deletion of this domain would result in no PIF complex in the ODV envelope. To address this question, we analyzed PIF complex formation in ODV envelopes by nondenaturing polyacrylamide gels and Western blot analysis as previously described (16) from the following viruses: AC83HA, TMΔ, CysΔ, 2NΔ, INM-SM-2NΔ, and the WT (AcMNPV-E2). The presence of the PIF complex was detected with anti-PIF1, anti-PIF2, or anti-HA antibodies. Blots probed with anti-PIF1 detected a band of ∼420 kDa corresponding to the intact PIF complex in the WT and AC83HA as expected. A faint band migrating at a lower molecular mass than the WT or AC83HA PIF complex was observed in TMΔ (TMΔ). No band was observed in CysΔ, 2NΔ, or INM-SM+2NΔ (Fig. 8). With anti-PIF2, the PIF complex was detected in both the WT and AC83HA but not in TMΔ, CysΔ, 2NΔ, or INM-SM+2NΔ. An identical PIF complex blot was probed with anti-HA antibodies, and similar results were obtained for the mutant viruses. As expected, the WT lane shows no band, since AC83 is not tagged with the HA epitope. The PIF complex band at ∼420 kDa, corresponding to intact PIF complex, was detected for AC83HA. A lower-molecular-weight band was detected for TMΔ, and no band was observed for CysΔ, 2NΔ, or INM-SM+2NΔ. These data show that the N-terminal region, specifically the cysteine-rich ZF domain, is essential for PIF complex formation in ODV. The TMΔ virus, which retains the cysteine-rich ZF domain, shows that the intact TM is not essential and that a partial PIF complex appears to form. The incomplete formation of the PIF complex would agree with the bioassay data, which showed that ODV from TMΔ was still infectious, but the LD50 was increased 10-fold. The absence of lower-molecular-mass bands in both blots, corresponding to either PIF1 or AC83, indicates the absence of noncomplexed PIF proteins in the ODV envelope. It is also possible that noncomplexed PIF proteins are being degraded.
FIG 8.
Native PAGE analysis of the ODV PIF complex of WT, AC83HA, TMΔ, CysΔ, 2NΔ, and INM-SM+2NΔ. ODV membrane proteins were purified and separated by native PAGE, followed by Western blotting with antibodies against PIF1, PIF2, or HA epitope as indicated below the panels. The blots showed a normal PIF complex in WT and AC83HA viruses. PIF complex formation was impaired in TMΔ and a lower-molecular-weight complex was observed. No PIF complex was observed in ODV from CysΔ, 2NΔ, or INM-SM+2NΔ, all of which contain deletions of the ZF domain.
Cellular localization of AC83 mutants.
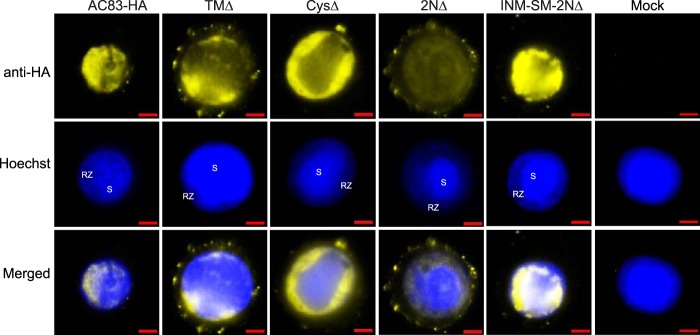
The results of previous studies (13, 17) have shown that AC83 is required for nucleocapsid assembly, which is essential for the production of both ODV and BV. This study showed that AC83 localizes to the nucleocapsid of both ODV and BV and was also inserted into the ODV envelope. ODV develop wholly in the nucleus, whereas BV nucleocapsids assemble in the nucleus, followed by budding from the plasma membrane. The results described above showed that loss of the ZF domain results in noninfectious ODV due to the loss of PIF complex assembly or recruitment, whereas the BVs that are produced remained infectious. We therefore compared by fluorescence microscopy the cellular localization of the AC83 proteins of AC83HA-, TMΔ-, CysΔ-, 2NΔ-, and INM-SM-2NΔ-infected cells (Fig. 9). Full-length AC83HA is found predominately in the nucleus, where it is distributed throughout the virogenic stroma where nucleocapsids form, but it concentrates to high levels in the ring zone on the periphery of the virogenic stroma where the ODVs develop and are occluded. Full-length AC83HA was barely detectable in the cytoplasm. In cells infected with TMΔ, which has the transmembrane domain removed from the INM-SM, AC83 remains predominately nuclear, with an accumulation in the ring zone similar to that of full-length AC83. However, with TMΔ, AC83 is also detectable in the cytoplasm, with foci at the plasma membrane. Deletion of the ZF (CysΔ) while retaining the INM-SM results in a distribution that appears to be the same as that of AC83HA. The virus 2NΔ, which deletes both the INM-SM and ZF domains, produces an AC83 protein that is both nuclear and cytoplasmic. In the nucleus, 2NΔ is in the virogenic stroma and ring zone and is evenly distributed without any concentration in the ring zone. In addition, with 2NΔ, AC83 forms a significant number of foci at the plasma membrane. INM-SM-2NΔ adds the INM-SM back to 2NΔ and AC83 reverts back to a predominately nuclear localization pattern with ring zone concentration and with very few foci at the plasma membrane. Overall, these results show that nuclear localization is retained if the AC83 NAD is retained, but concentration to the ring zone where ODV form requires the INM-SM. In the absence of the INM-SM signal, cytoplasmic localization of AC83 and the number of plasma membrane foci are increased.
FIG 9.
Localization of AC83 from AC83HA, TMΔ, CysΔ, 2NΔ, and INM-SM+2NΔ in T. ni BTI-Tn5B1-4 cells. BTI-Tn5B1-4 cells were infected at an MOI of 5 and fixed at 48 hpi. Fluorescence microscopy was used to examine AC83 localization (yellow), and the nucleus was stained with Hoechst 33342 (blue). Noninfected cells were used as a control (Mock). Arrows indicate AC83 foci at the plasma membrane. RZ, ring zone; S, virogenic stroma. Cells were immunostained with rabbit anti-HA monoclonal antibodies followed by goat anti-rabbit IgG antibodies conjugated with Alexa Fluor 680 (yellow). The scale bar is 5 μm.
DISCUSSION
Baculovirus ODV requires PIF proteins to bind to and interact with insect midgut epithelia to initiate a primary infection. The seven PIF proteins previously identified (PIF0 to PIF6) share common features, which include being expressed from a core gene, being essential for per os infectivity, being an ODV envelope protein that contains transmembrane domains, and that deletion of a pif gene does not impact BV production or systemic infection in vivo. The exception to this was the recently identified ac110, which was reported not to be a core gene (14, 27), but in this study we show that ac110 is the 38th core gene. This confirms that all pif genes are core genes and that it is correct to consider ac110 to be pif7. The main objective of this study was a reanalysis of the core AcMNPV protein AC83 to understand its functional association with the PIF complex. AC83 has many of the attributes of a PIF protein: it is expressed from a core gene, it is an ODV envelope protein that has an N-terminal transmembrane domain, and it is associated with the PIF complex. However, unlike PIF proteins, there have been contradictory reports about the location of AC83 within the virion (membrane and nucleocapsid), and deletion of ac83 results in no BV or ODV being produced due to the lack of nucleocapsid assembly. In addition, it was proposed that AC83 is required for passage through the peritrophic matrix due to a predicted CBD; however, chitin binding activity has not been demonstrated (2, 13, 17). Further analysis has resolved these issues, showing that AC83 has all the properties of a true PIF protein and should be considered PIF8, but with the unique property that it contains a domain required for nucleocapsid interaction and assembly.
The previously predicted AC83 CBD was reanalyzed, and due to a lack of essential amino acids required for chitin binding, this region could not be a CBD. The analysis did show, however, that the cysteine-rich region from amino acids 135 to 402 (Fig. 2 and 3) conforms strongly to a pattern of cysteine residues that is consistent with this region containing three ZF motifs and not the single motif previously reported for AC83 (13, 19). ZF domains have very different functions from CBDs, including being protein interaction domains (23). Based on this analysis, a new working model was proposed for AC83 function (Fig. 2B). The model predicts the AC83 ZF domain recruits or enables assembly of PIF proteins in the ODV membranes, the INM-SM directs AC83 to the nuclear ODV envelopes, and the NAD is required for nucleocapsid assembly.
To test our AC83 model, bacmids were constructed which expressed HA-tagged AC83 deletions from a genome backbone from which the WT ac83 had been deleted. Zhu et al. (13) had previously shown that N-terminal deletions up to amino acid 451 did not impact BV production. In addition, C-terminal deletions from amino acids 600 to 847 were able to produce BV but at significantly reduced levels. The conclusion from that study was that the region from amino acids 451 to 600 was required for nucleocapsid assembly, but this peptide was never expressed separately. Our constructs have refined the NAD in AC83 to amino acids 487 to 656, and only this region is required for BV production. Interestingly, the virus expressing only the NAD produced the highest titer of BV (Fig. 4, 2NΔ+CΔ), suggesting this virus is optimized for nucleocapsid egress. Retention of the C terminus, including the PR domain, in the absence of the INM-SM or ZF domains significantly reduces BV titers, suggesting this region can inhibit the egress pathway. Analysis of the NAD region does not reveal any predicted structures or motifs.
Previous analyses of AC83 and its homologs have variably reported that the virion location of this protein was only in the ODV envelope or that it was in both the envelope and the nucleocapsid (13, 19, 22). Our model predicted that AC83 should be located in both locations due to the NAD and INM-SM domains. Indeed, ODV fractionation revealed that full-length AC83 localized predominately to the ODV envelope, but a proportion also consistently localized to the nucleocapsid (Fig. 5A and B). The model also predicted that for viruses expressing only the AC83 NAD, the protein should localize solely to the nucleocapsid. As predicted, in ODV from the virus 1NΔ+CΔ, the NAD localized only to the nucleocapsid (Fig. 5A). These results indicate that AC83 is anchored in both the ODV envelope and nucleocapsid.
The AC83 ZF domain was predicted to be required for the assembly or recruiting of the PIF complex, and therefore deletion of this region should impact oral infectivity but not BV infectivity. This was confirmed with T. ni larval feeding bioassays of ODV and intrahemocoelic injection of BV. All of the constructs that contained deletions of the ZF domain were orally noninfectious, showing this domain was essential for per os infectivity. Deletion of the TM domain increased the LD90 10-fold, showing this domain impacts but is not essential for oral infectivity. Immunofluorescence images of infected midguts showed that deletion of the ZF domain renders the ODV unable to bind to and/or enter midgut cells of T. ni larvae. In contrast, intrahemocoelic injections of BV from all constructs were equally as infectious as AC83HA or WT virus.
The observation that ODV from viruses containing AC83 ZF deletions were unable to bind to insect midguts suggested that PIF complex formation in ODV envelopes was compromised. Therefore, the formation of the PIF complex was examined from the same ODV tested for per os infectivity. As expected, the PIF complex was detected in both WT and AC83HA ODV. TMΔ ODV, which is 10-fold less infectious than full-length AC83HA, contained a PIF complex, but it migrated at a lower molecular weight than WT or AC83HA, suggesting incomplete or inefficient PIF complex formation. No PIF complex was detectable in envelopes from ODV expressing proteins that do not contain the ZF domain. Previous analysis by Peng et al. (16) showed that deletion of pif4 results in loss of the 480-kDa PIF complex, similar to what is observed upon deletion of the AC83 ZF domain. However, despite the loss of the PIF complex in ODV envelopes of pif4-deleted viruses, PIF1 and PIF2 remained detectable by Western blotting as monomers or in lower-molecular-mass complexes in the tissue culture-derived ODV envelopes (16). In the current study, in the absence of the AC83 ZF domain PIF1 and PIF2 were not detectable in ODV envelopes. This suggests that if the AC83 ZF domain is deleted, the PIF complex is not being assembled or that PIF proteins are not being recruited to ODV envelopes. Recently it has been observed that when larva-derived ODVs of other PIF deletion viruses, which are disturbed in PIF complex formation, were isolated in the absence of protease inhibitors, PIF monomers also are not observed (B. Boogaard and M. van Oers, personal communication). Therefore, further studies will be required to determine if AC83 is required for assembly and/or for recruitment of the PIF complex in ODV envelopes.
Interestingly, when AC83 is isolated from ODV in the absence of protease inhibitors partial cleavage was observed, but only of the form that fractionates with the nucleocapsid (Fig. 5B). Previous studies on PIF0 (P74) have shown that protease cleavage of PIF0 is required for oral infectivity (25, 28). It remains to be determined if the AC83 cleavage plays any role in larval per os infectivity.
Proteomic analysis of various baculoviruses found that AC83 and its homologs were associated with ODV of AcMNPV (20), Culex nigripalpus NPV (29), Helicoverpa armigera NPV (26), and Pieris rapae granulovirus (30). Proteomic analysis of AcMNPV or Helicoverpa armigera NPV BV, however, did not identify AC83 (20, 26). In addition, previous studies also reported that AC83 was not found in BV of either AcMNPV or BmNPV when analyzed by Western blotting (13, 17). In contrast, OpMNPV P91 was found in both BV and ODV (22). In this study, we clearly show that AC83 is associated with BV, and unlike ODV, it is located almost entirely in the nucleocapsid fraction. We have also confirmed that AC83 is associated with BV by proteomic analysis of purified AcMNPV BV (S. Biswas and D. A. Theilmann, unpublished data). Since AC83 is essential for nucleocapsid formation, it is not surprising that it is present in BV. However, to permit BV egress, there must be some mechanism by which AC83 in BV nucleocapsids is prevented from interacting with ODV envelopes, which would sequester it in the nucleus. A possible mechanism includes modification of the AC83 INM-SM to prevent ODV envelope interaction. Alternatively, it could simply be that during the period of maximum BV production (18 to 24 hpi) there is a lack of ODV envelope synthesis and packaging of ODV nucleocapsids, which is known to occur late in infection (24 to 48 hpi) (31, 32).
The AC83 INM-SM was previously predicted and shown to function as a true nuclear envelope import signal similar to that reported for AcMNPV ODV-E66 (AC46) (13, 31). Our analysis showed that the AC83 INM-SM had a major impact on the cellular localization of AC83 proteins expressed in Sf9 cells. If the INM-SM domain was present, AC83 proteins were predominately in the nucleus but were significantly more concentrated in the ring zone where ODV are assembled than in the virogenic stroma. AC83 was also observed at very low levels in the cytoplasm and also formed foci at the plasma membrane which have not been previously described. Removal or disruption of the INM-SM resulted in AC83 proteins being evenly distributed in both the cytoplasm and nucleus and an increase in plasma membrane foci. Since AC83 is present in BV nucleocapsids, the plasma membrane foci could represent nucleocapsid budding sites, as have been previously identified with antibodies to GP64 (33). Overall, these data indicate that the INM-SM is essential for high-level accumulation in the ring zone, where ODV membranes concentrate and associate with nucleocapsids and ODV become occluded (31, 32). However, the data also show that AC83 is targeted to the nucleus in the absence INM-SM, indicating that there must be an alternative nuclear transport mechanism, which could be a nuclear localization signal (NLS), or that AC83 is transported to the nucleus in a complex with other proteins, as has been shown for helicase localization, which requires LEF3 (34). Analysis of AC83 did not identify any recognizable NLS signal. AC83 2NΔ+CΔ, which expresses only the NAD (amino acids 487 to 656), must be sufficient for nuclear transport, since it localizes to ODV nucleocapsids which only form within the nucleus, and Zhu et al. (13) have suggested that 597KRPK600 in this region represents an NLS. Concomitant with the submission of this study, Huang et al. (35) performed an in-depth analysis of the DNA sequences within NAD and identified a cis element that was required for budded virus production. The relationship between this cis element and the AC83 domain associated with nucleocapsid remains to be determined.
In conclusion, this study has reexamined AC83 and proposed and tested a model for its function. AC83 was shown to be a key PIF factor and therefore should be considered PIF8. Like all PIF proteins, it is expressed from a core gene is essential for per os infectivity. The caveat is that unlike all other PIF proteins identified, AC83 contains a 170-amino-acid NAD which is required for nucleocapsid assembly. The NAD may anchor AC83 to the nucleocapsid to enable the recruitment and/or assembly of the PIF complex in ODV envelopes. It remains to be determined what nucleocapsid proteins interact with this NAD to permit nucleocapsid development and if AC83 specifically interacts with a preformed PIF complex or individual PIF proteins. PIF proteins now represent 8/38 (29%) of the known baculovirus core genes and appear to be part of an essential infection mechanism that is utilized by baculoviruses from all genera and potentially other families of large nuclear-replicating invertebrate viruses that carry homologs of pif genes (36).
MATERIALS AND METHODS
Viruses and cell lines.
Spodoptera frugiperda clone 9 (Sf9) and Trichoplusia ni BTI-Tn5b1-4 cells were maintained in Grace's insect medium supplemented with l-glutamine, 3.33 g/liter lactalbumin hydrolysate, 3.33 g/liter yeastolate (Gibco Life Technologies), and 10% fetal bovine serum at 27°C. The AcMNPV strain used in this study was AcMNPV-E2. All AcMNPV bacmid constructs were developed using bMON14272 (37).
Construction of ac83 deletions and ac83 knockout (KO) and repair viruses.
To construct the ac83 deletion constructs, the ac83 gene was amplified by PCR using the primer pair 1639 (gcgctgcagggtttctacagtgcgttt) and 1640 (gcgtctagattaggcgtagtcgggcacgtcgtaggggtatacaatggaatcttcttgta) and ligated into the PstI and XbaI sites of the plasmid pBS+ (Stratagene). The amplified region included the ac83 late promoter, and the 3′ primer 1640 included the coding sequence for the HA epitope tag. The resulting plasmid (pAc83) was used to make all of the ac83 deletion constructs by a combination of inverse PCR and Gibson assembly (E2611S/L; NEB) by using the manufacturer's recommended protocol. All ac83 constructs were cloned into the pFAcT-Tnie1pA-GFP transposition vector using Gibson assembly (38). The pFAcT-Tnie1pA-GFP transfer vector, which contains polyhedrin under the control of its native promoter and gfp under the control of the OpMNPV ie1 promoter, was used to insert constructs into bacmids by Tn7-mediated transposition as previously described (38, 39).
An ac83 KO bacmid was made by replacing the ac83 coding sequence with an EM7-zeocin resistance cassette in the AcMNPV bacmid bMON14272 by homologous recombination as previously described (40). The EM7-zeocin resistance cassette was amplified using primers 2509 (gtacacgatcgcgttagagcaaatgatgaaaatcccaacgGGATCTCTGCAGCACGTGTT) and 2510 (caattcttcgggacacgccatgtattggccgtttttgacgtgcAGACATGATAAGATACATTGATGA) using the plasmid p2Zop2E as a template. Primer 2509 incorporates regions homologous to the flanking region of the 3′ end of ac83 (lowercase) and a region of the EM7 promoter-zeocin cassette (uppercase) (41). Primer 2509 incorporates regions homologous to the 5′ flanking region of ac83 (lowercase) and a region of the EM7 promoter-zeocin cassette (uppercase). The amplified EM7 promoter-zeocin resistance cassette was transformed into Escherichia coli BW25113 cells containing bMON14272 and the recombinase helper plasmid (pKD46) (40). A positive colony (ac83KO) was selected for kanamycin and zeocin resistance. Deletion of ac83 was confirmed by PCR. The bacmid ac83KO was transformed into E. coli DH10B cells containing the helper plasmid (pMON7124) which encodes the Tn7 transposase (37). Repair bacmids containing ac83 deletions were made by transforming them individually into DH10B cells that contained ac83KO and pMON7124.
To produce BV stocks, Sf9 cells were transfected using 1 μg of each repair bacmid and Lipofectin (42). BV supernatants were harvested at 5 days posttransfection and were used to infect 2.5 × 107 cells at an estimated multiplicity of infection (MOI) of 1. After 5 days at 27°C, the BV supernatants were collected. Viruses were named based on the AC83 product produced, namely, AC83-HA, TMΔ, CysΔ, 1NΔ, 2NΔ, 1NΔ+CΔ, 2NΔ+CΔ, 1NΔ+CΔ+PR, 2NΔ+CΔ+PR, INM-SM-2NΔ, INM-SM-2NΔ+CΔ+PR, and INM-SM-2NΔ+CΔ. Fifty percent tissue culture infective dose (TCID50) endpoint dilution was performed in duplicate to estimate viral titer by infecting Sf9 cells in 96-well microtiter plates (43).
OB production in T. ni larvae.
OBs were produced in T. ni 4th-instar larvae by injecting BV via hemocoelic injection of 100 TCID50 units/larva. OBs were purified from larval cadavers as previously described (44).
Extraction of PIF complex and native gel electrophoresis.
PIF protein complex was extracted as previously described, with minor modifications (16). Briefly, purified OBs (2 × 108) were pelleted by centrifugation at 3,500 × g for 5 min and incubated in 2 ml dilute alkaline saline (DAS; 0.1 M Na2CO3, 5 mM NaCl) at room temperature until dissolved, which was confirmed by phase-contrast microscopy (ca. 20 min). The suspension was centrifuged at 1,500 × g for 2 min to remove debris, and the supernatant consisting of ODVs was collected. ODVs were pelleted by centrifuging the suspensions at 20,600 × g for 25 min at 4°C. Pelleted ODVs were resuspended in 100 μl extraction buffer (6.25 mM Tris, 37.5 mM NaCl, 0.5% Triton X-100, pH 7.2) and sonicated briefly. The suspension was incubated overnight at 4°C with gentle rotation, and the ODV membrane proteins were collected in the supernatant following centrifugation at 20,600 × g for 20 min at 4°C. The ODV membrane preparation (20 μl of each sample) was mixed with native gel sample buffer (62.5 mM Tris-HCl, pH 6.8, 40% glycerol, and 0.01% bromophenol blue), and the protein complexes were separated on a precast 4 to 15% polyacrylamide gradient gel (Bio-Rad) in nondenaturing running buffer (25 mM Tris, 192 mM glycine). Separated protein bands were transferred onto polyvinylidene fluoride (PVDF) membranes for Western blot analysis.
Virion localization of AC83 and AC83 mutants.
ODV and BV were purified and the nucleocapsid and envelope fractions were isolated according to the procedure previously described by Braunagel and Summers (3).
Western blot analysis.
Proteins were separated by nondenaturing PAGE (described above) (45) using a Bio-Rad semidry Trans-Blot Turbo electroblotting system onto PVDF membranes (Millipore) using the manufacturer's protocols. Proteins separated by SDS-PAGE were performed using Bio-Rad Any kD 10-well TGX gels at 100 to 200 V for 30 to 60 min and transferred to PVDF membranes by electroblotting overnight at 30 V at 4°C. Blots were probed with mouse anti-HA monoclonal antibodies (MMS-101R-1000; Covance) diluted 1:2,000, anti-PIF1 or anti-PIF2 rat polyclonal antibodies (15) diluted 1:2,000, or rabbit anti-VP39 polyclonal primary antibodies diluted 1:5,000. Bound antibodies were detected with a secondary antibody conjugated to horseradish peroxidase, namely, goat anti-mouse IgG (no. 115-035-003; 1:15,000; Jackson ImmunoResearch Laboratory Inc.), goat anti-rat IgG (112-035-003; 1:15,000; Jackson ImmunoResearch Laboratory Inc.), or goat anti-rabbit IgG (111-035-003; 1:15,000; Jackson ImmunoResearch Laboratory Inc.). ODV HA-tagged proteins were detected using a primary rat polyclonal anti-HA antibody linked to horseradish peroxidase (11867423001; Sigma) (1:1,000). Bound peroxidase-linked antibodies were detected with Clarity Western ECL blotting substrate (Bio-Rad), developed according to the manufacturer's protocol, and imaged using a ChemiDoc MP system and Image Lab software (Bio-Rad).
Bioassays.
Per os infectivity of viruses was determined by leaf disc bioassays as previously described (44).
Localization of AC83 and AC83 deletion mutants in BTI-Tn5B1-4 cells.
T. ni BTI-Tn5B1-4 cells were seeded at 4 × 105 cells per ml into each well of a 12-well cell culture plate containing a sterilized 18-mm circular glass coverslip in TC100 cell culture medium supplemented with 10% fetal bovine serum and gentamicin (12.5 μg/ml). The cells were allowed to adhere to the coverslip for 4 h at 27°C. Cells were infected with BV at an MOI of 5 (triplicate wells for each virus). Uninfected wells were used as cell autofluorescence controls, 3 wells were used as control cells for detecting autofluorescence in virus-infected cells, and 3 additional virus-infected wells were used as fluorescence controls for secondary antibody nonspecific binding. At 48 hpi, medium was removed and the cell layer was washed twice with sterile phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4), and cells were fixed with 4% (vol/vol) formaldehyde in PBS for 20 min at room temperature and then washed 3 times with PBS. The cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature, washed 3 times with PBS, and blocked with 10% goat serum in PBS for 1 h at room temperature. All the wells, except the infection autofluorescence control and secondary antibody nonspecific binding controls, were probed with mouse monoclonal anti-HA antibodies (Abcam) diluted 1:400 in blocking reagent. After incubating the plates for 2 h at room temperature, cells were washed 3 times for 10 min with 1% goat serum in PBS. All of the wells except for the cell autofluorescence and infection autofluorescence controls were exposed to goat anti-mouse IgG conjugated with Alexa Fluor 680 secondary antibodies (Abcam) diluted 1:500 in blocking reagent. Plates were wrapped in aluminum foil and incubated at 4°C overnight, followed by staining with 2 μg/ml Hoechst 33342 (Invitrogen) in 1% goat serum and incubation for 10 min at room temperature. Cells were washed 3× with 1% goat serum for 10 min and examined with an Imager Z1 optical fluorescence microscope (Carl Zeiss, Canada). Images were captured using an Axiocam HRm camera and processed with AxioVision software (Carl Zeiss, Canada).
Analysis of BV production.
BV production at 24 and 48 hpi was determined in Sf9 cells. Sf9 cells were seeded into 6-well tissue culture plates at a density of 1.0 × 106 cells per well and were allowed to adhere to the well surface for 1 h. Cells were infected with BV at an MOI of 10 TCID50/cell and allowed to adsorb with rocking for 1 h. Cells were washed with 1× PBS and then overlaid with 1.5 ml TC100 medium. At 24 and 48 hpi, supernatants containing BV were collected into 2-ml microtubes. Free cells were pelleted by centrifugation at 3,000 × g for 10 min at 4°C, and the supernatants containing the BV were stored at 4°C.
Relative BV titers were determined by droplet digital quantitative PCR (ddPCR) using a modification of a quantitative PCR (qPCR) protocol (46). Droplets for ddPCR were generated and read using the Bio-Rad QX200 system. For each virus, 20 μl of BV supernatant was digested with Proteinase K (5 mM Tris · Cl, pH 8.0, 0.5 mM EDTA, 0.5% SDS, Proteinase K [80 μg/ml]) overnight at 50°C. Reaction mixtures were heated at 100°C for 15 min to inactivate the Proteinase K. From the Proteinase K-digested material 1 μl was diluted 100-fold in 99 μl distilled H2O (dH2O). PCRs were performed using a Bio-Rad MyCycler using the following reaction mix: 5.5 μl of the diluted Proteinase K-treated sample, 11 μl of Bio-Rad 2× QX200 EvaGreen super mix (no. 186-4034), 1.5 μl of primer 850 (5′-TTTGGCAAGGGAACTTTGTC-3′) (2.2 μM), 1.5 μl of primer 851 (5′-ACAAACCTGGCAGGAGAGAG-3′) (2.2 μM), and 2.5 μl dH2O. Thermocycler conditions were 95°C for 5 min (1×); 95°C for 30 s and 58°C for 80 s (44×); 4°C for 5 min (ramp 2°C/s); 95°C for 5 min (ramp 2°C/s); and cooling to 4°C.
Homology modeling.
The theoretical structure of the AC83 cysteine-rich region of AC83 (residues 131 to 403) was investigated using the SWISS-MODEL server (swissmodel.expasy.org) (47–49). Using the fully automated option within SWISS-MODEL, structural templates were identified and a preliminary model for the aligned residues of AC83 was produced. The template selected as the best model was chain A from the NMR structure of tachycitin (PDB entry 1DQC), a 72-residue antimicrobial protein with chitin binding function (50), which aligned from threonine 142 to arginine 180. Modeling of putative ZF domains within AC83 was attempted using the fully automated mode of SWISS-MODEL. Structural templates for these putative domains for homology modeling were not available.
ACKNOWLEDGMENTS
This study was supported in part by funding from the Natural Sciences and Engineering Research Council (D.A.T.). Funding was also provided by Agriculture and Agri-Food Canada (D.A.T, D.D.H., B.C.D., and M.A.E).
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JVI.02110-16.
REFERENCES
- 1.Herniou EA, Olszewski JA, Cory JS, O'Reilly DR. 2003. The genome sequence and evolution of baculoviruses. Annu Rev Entomol 48:211–234. doi: 10.1146/annurev.ento.48.091801.112756. [DOI] [PubMed] [Google Scholar]
- 2.Rohrmann GF. 2013. Baculovirus molecular biology. National Center for Biotechnology Information, Bethesda, MD. [PubMed] [Google Scholar]
- 3.Braunagel SC, Summers MD. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 4.Haas-Stapleton EJ, Washburn JO, Volkman LE. 2004. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. J Virol 78:6786–6791. doi: 10.1128/JVI.78.13.6786-6791.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton HM, Burand JP. 1993. Saturable attachment sites for polyhedron-derived baculovirus on insect cells and evidence for entry via direct membrane fusion. J Virol 67:1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faulkner P, Kuzio J, Williams GV, Wilson JA. 1997. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J Gen Virol 78(Part 12):3091–3100. doi: 10.1099/0022-1317-78-12-3091. [DOI] [PubMed] [Google Scholar]
- 7.Kikhno I, Gutierrez S, Croizier L, Croizier G, Ferber ML. 2002. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J Gen Virol 83:3013–3022. doi: 10.1099/0022-1317-83-12-3013. [DOI] [PubMed] [Google Scholar]
- 8.Pijlman GP, Pruijssers AJ, Vlak JM. 2003. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J Gen Virol 84:2041–2049. doi: 10.1099/vir.0.19133-0. [DOI] [PubMed] [Google Scholar]
- 9.Fang M, Nie Y, Harris S, Erlandson MA, Theilmann DA. 2009. Autographa californica multiple nucleopolyhedrovirus core gene ac96 encodes a per os infectivity factor (PIF-4). J Virol 83:12569–12578. doi: 10.1128/JVI.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J Virol 79:15258–15264. doi: 10.1128/JVI.79.24.15258-15264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison RL, Sparks WO, Bonning BC. 2010. Autographa californica multiple nucleopolyhedrovirus ODV-E56 envelope protein is required for oral infectivity and can be substituted functionally by Rachiplusia ou multiple nucleopolyhedrovirus ODV-E56. J Gen Virol 91:1173–1182. doi: 10.1099/vir.0.017160-0. [DOI] [PubMed] [Google Scholar]
- 12.Nie Y, Fang M, Erlandson MA, Theilmann DA. 2012. Analysis of the autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication. J Virol 86:3985–3994. doi: 10.1128/JVI.06849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu S, Wang W, Wang Y, Yuan M, Yang K. 2013. The baculovirus core gene ac83 is required for nucleocapsid assembly and per os infectivity of Autographa californica nucleopolyhedrovirus. J Virol 87:10573–10586. doi: 10.1128/JVI.01207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiantao L, Zhu L, Zhang S, Deng Z, Huang Z, Yuan M, Wu W, Yang K. 2016. The Autographa californica multiple nucleopolyhedrovirus ac110 gene encodes a new per os infectivity factor. Virus Res 221:30–37. doi: 10.1016/j.virusres.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Peng K, van Oers MM, Hu Z, van Lent JW, Vlak JM. 2010. Baculovirus per os infectivity factors form a complex on the surface of occlusion-derived virus. J Virol 84:9497–9504. doi: 10.1128/JVI.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng K, van Lent JW, Boeren S, Fang M, Theilmann DA, Erlandson MA, Vlak JM, van Oers MM. 2012. Characterization of novel components of the baculovirus per os infectivity factor complex. J Virol 86:4981–4988. doi: 10.1128/JVI.06801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang X, Shen Y, Yang R, Chen L, Hu X, Wu X. 2013. Bombyx mori nucleopolyhedrovirus BmP95 plays an essential role in budded virus production and nucleocapsid assembly. J Gen Virol 94:1669–1679. doi: 10.1099/vir.0.050583-0. [DOI] [PubMed] [Google Scholar]
- 18.Ono C, Kamagata T, Taka H, Sahara K, Asano S, Bando H. 2012. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res 165:197–206. doi: 10.1016/j.virusres.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Lu M, Swevers L, Iatrou K. 1998. The p95 gene of Bombyx mori nuclear polyhedrosis virus: temporal expression and functional properties. J Virol 72:4789–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braunagel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc Natl Acad Sci U S A 100:9797–9802. doi: 10.1073/pnas.1733972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Deng F, Hou D, Zhao Y, Guo L, Wang H, Hu Z. 2010. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J Virol 84:7233–7242. doi: 10.1128/JVI.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell RL, Rohrmann GF. 1997. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology 233:210–223. doi: 10.1006/viro.1997.8599. [DOI] [PubMed] [Google Scholar]
- 23.Krishna SS, Majumdar I, Grishin NV. 2003. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res 31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Christensen RG, Bell HA, Goodwin M, Patel RY, Pandey M, Enuameh MS, Rayla AL, Zhu C, Thibodeau-Beganny S, Brodsky MH, Joung JK, Wolfe SA, Stormo GD. 2014. An improved predictive recognition model for Cys(2)-His(2) zinc finger proteins. Nucleic Acids Res 42:4800–4812. doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng K, van Lent JW, Vlak JM, Hu Z, van Oers MM. 2011. In situ cleavage of baculovirus occlusion-derived virus receptor binding protein P74 in the peroral infectivity complex. J Virol 85:10710–10718. doi: 10.1128/JVI.05110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou D, Zhang L, Deng F, Fang W, Wang R, Liu X, Guo L, Rayner S, Chen X, Wang H, Hu Z. 2013. Comparative proteomics reveal fundamental structural and functional differences between the two progeny phenotypes of a baculovirus. J Virol 87:829–839. doi: 10.1128/JVI.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garavaglia MJ, Miele SA, Iserte JA, Belaich MN, Ghiringhelli PD. 2012. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J Virol 86:12069–12079. doi: 10.1128/JVI.01873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slack JM, Lawrence SD, Krell PJ, Arif BM. 2008. Trypsin cleavage of the baculovirus occlusion-derived virus attachment protein P74 is prerequisite in per os infection. J Gen Virol 89:2388–2397. doi: 10.1099/vir.0.2008/002543-0. [DOI] [PubMed] [Google Scholar]
- 29.Perera O, Green TB, Stevens SM Jr, White S, Becnel JJ. 2007. Proteins associated with Culex nigripalpus nucleopolyhedrovirus occluded virions. J Virol 81:4585–4590. doi: 10.1128/JVI.02391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XF, Zhang BQ, Xu HJ, Cui YJ, Xu YP, Zhang MJ, Han YS, Lee YS, Bao YY, Zhang CX. 2011. ODV-associated proteins of the Pieris rapae granulovirus. J Proteome Res 10:2817–2827. doi: 10.1021/pr2000804. [DOI] [PubMed] [Google Scholar]
- 31.Braunagel SC, Summers MD. 2007. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr Drug Targets 8:1084–1095. doi: 10.2174/138945007782151315. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Li K, Tang P, Li Y, Zhou Q, Yang K, Zhang Q. 2015. Three-dimensional visualization of the Autographa californica multiple nucleopolyhedrovirus occlusion-derived virion envelopment process gives new clues as to its mechanism. Virology 476:298–303. doi: 10.1016/j.virol.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 33.de Jong J, Theilmann DA, Arif BM, Krell PJ. 2011. Immediate-early protein ME53 forms foci and colocalizes with GP64 and the major capsid protein VP39 at the cell membranes of Autographa californica multiple nucleopolyhedrovirus-infected cells. J Virol 85:9696–9707. doi: 10.1128/JVI.00833-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Carstens EB. 2005. Identification of domains in Autographa californica multiple nucleopolyhedrovirus late expression factor 3 required for nuclear transport of P143. J Virol 79:10915–10922. doi: 10.1128/JVI.79.17.10915-10922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Pan M, Zhu S, Zhang H, Wu W, Yuan M, Yang K. 2016. The Autographa californica nucleopolyhedrovirus ac83 gene contains a cis-acting element that is essential for nucleocapsid assembly. J Virol 91:e02110-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ZH, Olafsson R, Ingram P, Li Q, Qin Y, Witte RS. 2011. Four-dimensional ultrasound current source density imaging of a dipole field. Appl Physics Lett 99:113701–1137013. doi: 10.1063/1.3632034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol 67:4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nie Y, Fang M, Theilmann DA. 2009. AcMNPV AC16 (DA26, BV/ODV-E26) regulates the levels of IE0 and IE1 and binds to both proteins via a domain located within the acidic transcriptional activation domain. Virology 385:484–495. doi: 10.1016/j.virol.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 39.Dai X, Stewart TM, Pathakamuri JA, Li Q, Theilmann DA. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J Virol 78:9633–9644. doi: 10.1128/JVI.78.18.9633-9644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeifer TA, Hegedus DD, Grigliatti TA, Theilmann DA. 1997. Baculovirus immediate-early promoter-mediated expression of the Zeocin resistance gene for use as a dominant selectable marker in dipteran and lepidopteran insect cell lines. Gene 188:183–190. doi: 10.1016/S0378-1119(96)00756-1. [DOI] [PubMed] [Google Scholar]
- 42.Campbell MJ. 1995. Lipofection reagents prepared by a simple ethanol injection technique. Biotechniques 18:1027–1032. [PubMed] [Google Scholar]
- 43.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 44.Erlandson M, Newhouse S, Moore K, Janmaat A, Myers J, Theilmann D. 2007. Characterization of baculovirus isolates from Trichoplusia ni populations from vegetable greenhouses. Biol Control 41:256–263. doi: 10.1016/j.biocontrol.2007.01.011. [DOI] [Google Scholar]
- 45.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Sokal N, Nie Y, Willis LG, Yamagishi J, Blissard GW, Rheault MR, Theilmann DA. 2014. Defining the roles of the baculovirus regulatory proteins IE0 and IE1 in genome replication and early gene transactivation. Virology 468-470:160–171. [DOI] [PubMed] [Google Scholar]
- 47.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 48.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13. [DOI] [PubMed] [Google Scholar]
- 50.Suetake T, Tsuda S, Kawabata S, Miura K, Iwanaga S, Hikichi K, Nitta K, Kawano K. 2000. Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J Biol Chem 275:17929–17932. doi: 10.1074/jbc.C000184200. [DOI] [PubMed] [Google Scholar]
- 51.Toprak U, Baldwin D, Erlandson M, Gillott C, Harris S, Hegedus DD. 2010. Expression patterns of genes encoding proteins with peritrophin A domains and protein localization in Mamestra configurata. J Insect Physiol 56:1711–1720. doi: 10.1016/j.jinsphys.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Tetreau G, Dittmer NT, Cao X, Agrawal S, Chen YR, Muthukrishnan S, Haobo J, Blissard GW, Kanost MR, Wang P. 2015. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem Mol Biol 62:127–141. doi: 10.1016/j.ibmb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]