ABSTRACT
Dengue virus (DENV) is a member of the genus Flavivirus and can cause severe febrile illness. Here, we show that FLJ11286, which we refer to as IRAV, is induced by DENV in an interferon-dependent manner, displays antiviral activity against DENV, and localizes to the DENV replication complex. IRAV is an RNA binding protein and localizes to cytoplasmic processing bodies (P bodies) in uninfected cells, where it interacts with the MOV10 RISC complex RNA helicase, suggesting a role for IRAV in the processing of viral RNA. After DENV infection, IRAV, along with MOV10 and Xrn1, localizes to the DENV replication complex and associates with DENV proteins. Depletion of IRAV or MOV10 results in an increase in viral RNA. These data serve to characterize an interferon-stimulated gene with antiviral activity against DENV, as well as to propose a mechanism of activity involving the processing of viral RNA.
IMPORTANCE Dengue virus, a member of the family Flaviviridae, can result in a life-threatening illness and has a significant impact on global health. Dengue virus has been shown to be particularly sensitive to the effects of type I interferon; however, little is known about the mechanisms by which interferon-stimulated genes function to inhibit viral replication. A better understanding of the interferon-mediated antiviral response to dengue virus may aid in the development of novel therapeutics. Here, we examine the influence of the interferon-stimulated gene IRAV (FLJ11286) on dengue virus replication. We show that IRAV associates with P bodies in uninfected cells and with the dengue virus replication complex after infection. IRAV also interacts with MOV10, depletion of which is associated with increased viral replication. Our results provide insight into a newly identified antiviral gene, as well as broadening our understanding of the innate immune response to dengue virus infection.
KEYWORDS: IRAV, FLJ11286, C19orf66, UPF0515, dengue, flavivirus, interferon, MOV10, P body
INTRODUCTION
Dengue virus (DENV), a member of the genus Flavivirus, is a positive-sense RNA virus comprised of a nucleoprotein core surrounded by a host-derived membrane. DENV is transmitted through the bite of a mosquito vector and is the etiologic agent of a spectrum of illnesses that can range from mild fever to the potentially fatal dengue hemorrhagic fever/dengue shock syndrome. Due to geographic expansion of its insect vector, as well as increased travel and urbanization, dengue virus is of increasing importance to public health. Dengue virus infects an estimated 390 million people each year, with more than 2.5 billion people living in regions where they are at risk of dengue virus transmission (1–4).
Activation of interferon (IFN) signaling pathways results in the upregulation of hundreds of interferon-stimulated genes (ISGs) (5–7). Interferon-stimulated genes encoding viperin, TRAIL, IFITM1, IFITM2, IFITM3, ISG20, and TRIM56 have all been shown to inhibit DENV replication through a variety of mechanisms (8–12). In addition, a number of ISGs, including IFIT2, IFIT1, ISG15, TRIM22, and TRIM79α genes, have been found to have activity against other flaviviruses (13–18). High-throughput screens have implicated a number of other ISGs in inhibition of flavivirus replication (6, 19).
Previously, we identified FLJ11286 as one of a number of genes that were upregulated in Daudi cells in response to treatment with IFN (20) and showed that they displayed antiviral activity against encephalomyocarditis virus (EMCV) (21). FLJ11286 has also been shown by our laboratory and others to have antiviral activity against DENV (22, 23). FLJ11286, which we refer to here as IRAV (interferon-regulated antiviral gene) (also annotated as C19orf66, UPF0515, or RyDEN), encodes a protein 291 amino acids (aa) in length with a calculated molecular mass of 33.1 kDa. Analysis of published microarray data suggests that IRAV (FLJ11286) is upregulated in response to type I and type II IFNs (6, 20, 24–26). IRAV (FLJ11286) has been shown in microarray screens to be upregulated in response to the yellow fever virus vaccine (27), as well as after infection with a number of different pathogens, including adenovirus (28), influenza virus (29), Lassa virus (30), and ebola and Marburg viruses (31), as well as after infection with human herpesvirus 8 and human herpesvirus 1 (32, 33). In addition, proteomic analysis of the hepatitis C virus (HCV) interactome identified IRAV (FLJ11286) as one of 214 human proteins interacting with the HCV NS3 protein (34), suggesting a role for IRAV in the host pathogen response.
Here, we show that IRAV is upregulated in response to DENV infection in an IFN-dependent manner. Upregulation of IRAV in response to IFN-β treatment can be blocked by disrupting the canonical ISGF3 pathway. CRISPR/Cas9-mediated knockout of IRAV resulted in increased titers of DENV, as well as of EMCV. We also demonstrate that IRAV associates with DENV proteins and localizes to the viral replication complex. IRAV is an RNA binding protein and localizes to P bodies in uninfected cells. IRAV also associates with the host RNA binding proteins UPF1 and HuR (ELAV1) and interacts with MOV10 (a RISC complex RNA helicase), suggesting a role for IRAV in processing or stability of RNA. Furthermore, we propose a mechanism of action for IRAV that utilizes intrinsic RNA decay pathways. These pathways have been shown to be of increasing importance to the life cycles of multiple viruses, as well as in an array of cellular processes.
RESULTS
IRAV is upregulated after dengue virus infection in an interferon-dependent manner.
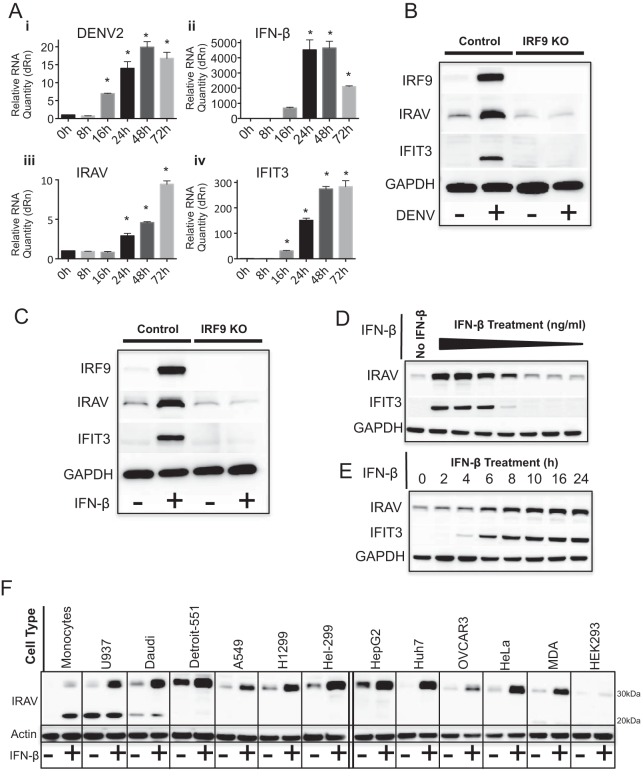
To determine if IRAV is upregulated in response to DENV infection, A549 cells were infected with DENV for 72 h. Quantitative real-time PCR (qRT-PCR) analysis showed upregulation of IRAV starting at 24 h postinfection, corresponding to increased expression of DENV RNA, IFN-β, and ISGs, including IFIT3 (Fig. 1A). To determine if IRAV is regulated through the canonical IFN (ISGF3) signaling pathway, IRF9 knockouts (KO) were generated in A549 cells using CRISPR/Cas9. Knockout of IRF9 resulted in decreased expression of IRAV after DENV infection (Fig. 1B), as well as after IFN-β treatment (Fig. 1C), indicating that the canonical ISGF3 pathway plays a role in induction of IRAV.
FIG 1.
IRAV is an interferon-stimulated gene. (A) qRT-PCR analysis of RNA expression in A549 cells after DENV infection at 0, 8, 16, 24, 48, and 72 h postinfection. Cells were infected with DENV at an MOI of 0.1 for 1 h, and samples were collected at time zero and the indicated time points. The samples were analyzed by qRT-PCR for DENV RNA (i), as well as for expression of IFN-β (ii), IRAV (iii), and the ISG IFIT3 (iv). The samples were normalized to the HPRT housekeeping gene, and the change in expression was calculated relative to time zero. The error bars represent standard deviations. *, P < 0.05. (B) Western blot analysis of IRAV expression in control A549 cells or IRF9 KO cells after DENV infection (72 h). Uninfected (−) and DENV-infected (+) A549 or IRF9 KO cells were analyzed by Western blotting for IRF9, IRAV, or IFIT3. GAPDH was used as a loading control. (C) Western blot analysis of IRAV expression in control A549 or IRF9 KO cells after treatment with IFN-β. The cells were either left untreated (−) or treated with IFN-β (+) for 16 h, followed by Western blot analysis for expression of IRF9, IRAV, or IFIT3. GAPDH was used as a loading control. (D) Western blot analysis of IRAV expression in HeLa cells after treatment with 10-fold dilutions of IFN-β (30 to 0.00003 ng/ml) for 16 h. Samples were examined by Western blotting for expression of IRAV and IFIT3. GAPDH was used as a loading control. (E) Western blot analysis of IRAV expression in HeLa cells after treatment with IFN-β (3 ng/ml) at the indicated time points. Samples were examined by Western blotting for expression of IRAV and IFIT3. GAPDH was used as a loading control. (F) Western blot analysis of IRAV expression in various cell lines. The indicated cell lines were either left untreated (−) or treated with IFN-β (+) (10 ng/ml) for 16 h, followed by Western blot analysis for IRAV. Actin was used as a loading control.
To further characterize IRAV induction in response to IFN, HeLa cells were treated with various concentrations of IFN-β for 16 h or were treated with IFN-β at a concentration of 3 ng/ml and collected at the indicated time points. Samples were analyzed by Western blotting for expression of IRAV and IFIT3. Analysis showed IRAV to be detectable in unstimulated HeLa cells and to be upregulated in response to treatment with IFN-β in both dose-dependent (Fig. 1D) and time-dependent (Fig. 1E) manners. To determine if IRAV was expressed in different cell lines, cells were left untreated or treated with IFN-β (10 ng/ml) for 16 h, followed by Western blot analysis for IRAV. The results showed that IRAV was upregulated in all the tested cell lines, with the exception of HEK293 cells, which produced only low levels of IRAV. In addition, it was observed that monocytes and U937 and Daudi cells produced an additional low-molecular-mass band of approximately 26 kDa that matched the predicted molecular mass of an alternate IRAV isoform (Fig. 1F).
CRISPR-mediated knockout of IRAV results in increased titers of DENV and EMCV.
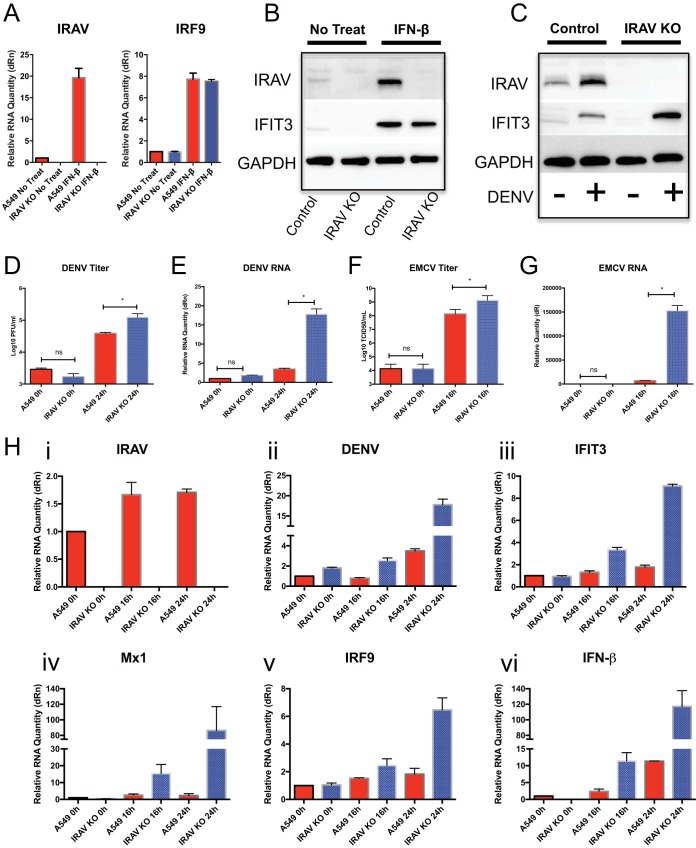
The CRISPR/Cas9 system was used to generate A549 IRAV KO cells. Treatment of A549 cells with IFN-β resulted in upregulation of IRAV, as well as of other ISGs. However, in the KO cells, IRAV was not detected at either the RNA (Fig. 2A) or protein (Fig. 2B) level, while expression of other ISGs remained unaffected. Infection of A549 or IRAV KO cells with DENV (Fig. 2C) resulted in increased titers of DENV (Fig. 2D), as well as significant increases in DENV RNA relative to control cells (Fig. 2E). Similar effects were observed after infection with EMCV (Fig. 2F and G). When DENV-infected IRAV KO cells were examined for expression of other genes (Fig. 2H), we observed increased expression of the IFIT3, Mx1, IRF9, and IFN-β ISGs in IRAV KO cells relative to the control A549 cells. These results suggest that IRAV directly affects DENV replication rather than exerting its effects via regulation of other ISGs.
FIG 2.
Knockout of IRAV results in enhanced replication of DENV and EMCV. (A) A549 cells or IRAV KO cells were left untreated or treated with IFN-β for 16 h, followed by qRT-PCR analysis of RNA expression of IRAV or IRF9. Samples were normalized to the HPRT housekeeping gene, and the change in expression was calculated relative to time zero. (B) Western blot analysis of A549 control or IRAV KO cells after IFN treatment. The cells were either left untreated or treated with IFN-β for 16 h, followed by Western blot analysis for IRAV or IFIT3. GAPDH was used as a loading control. (C) Western blot analysis of A549 control or IRAV KO cells after DENV infection. The cells were either left untreated (−) or infected with DENV for 72 h (+), followed by Western blotting for IRAV or IFIT3. GAPDH was used as a loading control. (D) A549 or IRAV KO cells were infected with DENV, and samples were collected at time zero and 24 h, followed by titration using the plaque assay method. (E) A549 or IRAV KO cells were infected with DENV, and samples were collected at time zero and 24 h, followed by qRT-PCR analysis of DENV RNA. (F) A549 or IRAV KO cells were infected with EMCV, and samples were collected at time zero and 16 h, followed by titration using the 50% tissue culture infective dose (TCID50) method. (G) A549 or IRAV KO cells were infected with EMCV, and samples were collected at time zero and 16 h, followed by qRT-PCR analysis of EMCV RNA. (H) qRT-PCR analysis of A549 or IRAV KO cells infected with DENV. Samples were collected at time zero and 16 h and 24 h postinfection and analyzed for IRAV (i) and DENV RNA (ii), as well as for expression of IFIT3 (iii), Mx1 (iv), IRF9 (v), and IFN-β (vi). Samples were normalized to the HPRT housekeeping gene, and the change in expression was calculated relative to time zero. The error bars represent standard deviations. *, P < 0.05; ns, not significant.
IRAV associates with the dengue virus replication complex.
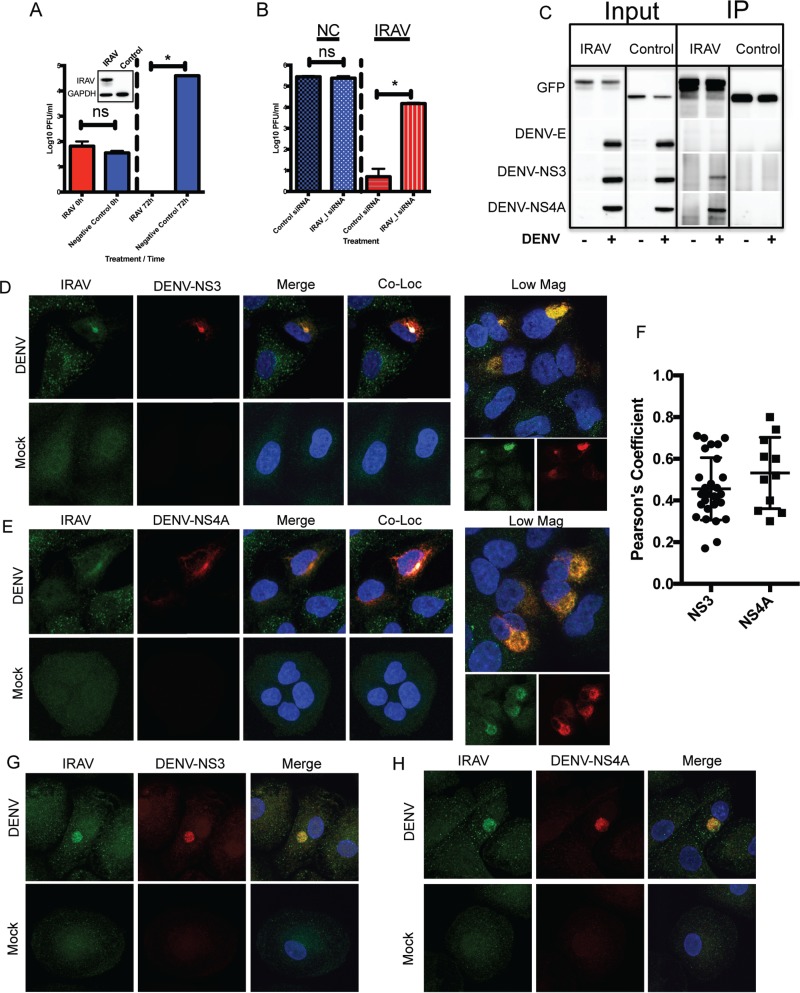
To determine if IRAV has a direct influence on virus replication, HEK293 cells were stably transfected with an expression vector encoding IRAV with an amino-terminal (N-terminal) green fluorescent protein (GFP) fusion or with a negative-control vector with an N-terminal GFP tag. Because IRAV is poorly expressed in HEK293 cells (Fig. 1F), they are an ideal cell line for overexpression experiments. Cells were infected with DENV (multiplicity of infection [MOI], 0.1), and supernatants were collected at time zero and at 72 h postinfection, and the titer was determined by plaque assay. While detectable in the negative-control group, no virus was detected in cells stably expressing IRAV (Fig. 3A). The IRAV-expressing HEK293 cells were also treated with a small interfering RNA (siRNA) targeting IRAV or with a negative-control siRNA. The cells were then infected with DENV at an MOI of 0.1 for 72 h. Virus titration showed a significant increase in virus titers in IRAV-expressing HEK293 cells after knockdown of IRAV (Fig. 3B), demonstrating that IRAV has a significant influence on DENV replication even in the absence of interferon treatment. Because IRAV was previously identified by mass spectrometry (MS) as an interaction partner for HCV NS3 (34), we examined whether IRAV was interacting with DENV proteins. Coimmunoprecipitation (co-IP) experiments were performed on DENV-infected cells using GFP-IRAV as bait and GFP-chloramphenicol acetyltransferase (CAT) as a negative control. Western blotting showed that the DENV protease/helicase NS3 and the replication complex-associated protein NS4A interacted with IRAV, while the DENV envelope (E) protein did not (Fig. 3C). Interactions between IRAV and NS3 were not affected by RNase A treatment (data not shown). To further substantiate our findings, colocalization experiments were performed in A549 cells. Here, A549 cells were infected with DENV serotype 2 (DENV-2) for 48 h, followed by fixation and immunostaining for IRAV and either DENV NS3 (Fig. 3D) or NS4A (Fig. 3E) protein. While IRAV was localized to small puncta in mock-infected cells, colocalization between IRAV and NS3 or NS4A was observed in perinuclear regions of DENV-infected cells (Fig. 3F). This suggests that IRAV relocalizes to the DENV replication complex after infection. In order to demonstrate association of IRAV with the DENV replication complex in primary human cells, monocyte-derived macrophages were infected with DENV-2 as described above. As shown for A549 cells, colocalization was observed between IRAV and DENV NS3 (Fig. 3G) or NS4A (Fig. 3H) in DENV-infected monocyte-derived macrophages.
FIG 3.
IRAV associates with the DENV replication complex. (A) HEK293 cells stably expressing IRAV or a negative control were infected with DENV at an MOI of 0.1, and samples were collected at time zero and 72 h postinfection, followed by titration using the plaque assay method. The inset represents expression of IRAV in transfected HEK293 cells compared to the negative control, as determined by Western blot analysis. GAPDH was used as a loading control. (B) HEK293 cells stably expressing IRAV or negative-control (NC) cells were treated with either negative-control siRNA or an siRNA specific to IRAV (IRAV_I). The cells were then infected with DENV for 72 h, followed by titration on Vero cells. (C) IRAV coimmunoprecipitates with DENV proteins. HEK293 cells were either left uninfected (−) or infected with DENV (+) for 48 h, followed by transfection with a plasmid expressing GFP-IRAV (IRAV) or GFP-CAT (Control) for an additional 48 h. The cell lysates were then collected, and co-IP experiments were performed using antibodies to GFP. Input and IP samples were then analyzed by Western blotting for the presence of DENV envelope (DENV-E), DENV NS3, or DENV NS4A protein. (D) Confocal microscopy of IRAV colocalized with replication complexes in DENV-infected or mock-infected A549 cells. Green, IRAV; red, DENV-NS3. The nucleus was stained with DAPI (blue). Colocalization between IRAV and DENV-NS3 is shown in white. Low Mag, a lower-magnification field showing a cluster of infected cells. (E) Confocal microscopy of IRAV colocalized with replication complexes in DENV-infected or mock-infected A549 cells. Green, IRAV; red, DENV NS4A. The nucleus was stained with DAPI (blue). Colocalization between IRAV and DENV NS4A is shown in white. (F) Colocalization coefficients of IRAV and DENV NS3 (NS3) or DENV NS4A (NS4A) in DENV-infected A549 cells as determined by Pearson's linear correlation coefficient. (G) Confocal microscopy of IRAV colocalized with replication complexes in DENV-infected or mock-infected monocyte-derived macrophages. Green, IRAV; red, DENV-NS3. The nucleus was stained with DAPI (blue). (H) Confocal microscopy of IRAV colocalized with replication complexes in DENV-infected or mock-infected monocyte-derived macrophages. Green, IRAV; red, DENV NS4A. The nucleus was stained with DAPI (blue). The error bars represent standard deviations.
IRAV associates with host RNA binding proteins.
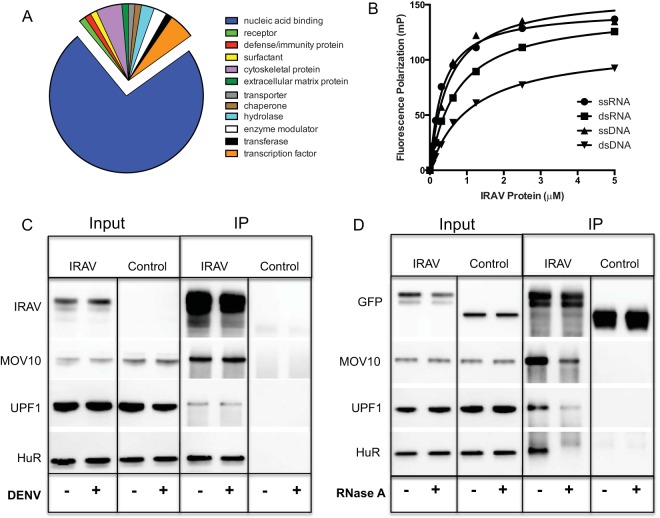
To determine if IRAV interacted with host proteins in DENV-infected HEK293 cells, co-IP experiments were performed using overexpressed IRAV as bait. The immunoprecipitated proteins were then analyzed by MS. Gene ontology (GO) analysis was performed using the PANTHER classification system (http://pantherdb.org) (35) on MS hits with a spectral count of >50. Ontology derived based on the protein class revealed a strong preference (74%) for interactions with nucleic acid binding proteins (Fig. 4A). To characterize the nucleic acid binding properties of IRAV, fluorescence polarization (FP) assays were performed using recombinant IRAV (with an amino-terminal maltose binding protein [MBP] tag and a carboxy-terminal truncation of 18 amino acids) and synthetic 20-mer nucleic acids. Here, dilutions of IRAV were incubated with either synthetic single-stranded (ssRNA) or double-stranded (dsRNA) RNA or single- or double-stranded DNA containing complementary nucleic acid sequences. FP showed IRAV to be a nucleic acid binding protein with a higher affinity for ssRNA and ssDNA than for dsDNA (Fig. 4B).
FIG 4.
IRAV interacts with RNA binding proteins. (A) Gene ontology analysis of IRAV interaction partners as identified by co-IP of GFP-IRAV from DENV-infected HEK293 cells, followed by MS analysis. Gene ontology analysis was performed using the PANTHER classification system (http://www.pantherdb.org), based on the protein class. (B) FP assays were performed using various concentrations of recombinant IRAV and FAM-labeled 20-mer single-stranded or double-stranded DNA or RNA oligomers. Samples were run in triplicate, and FP was measured on a Hidex sense microplate reader (Turku, Finland). (C) Co-IP experiments performed on HEK293 cells left untreated (−) or infected with DENV (+). The cells were transfected with expression vectors for GFP-IRAV (IRAV) or GFP-CAT (Control), followed by IP with an antibody for GFP. Western blotting was performed for IRAV putative interaction partners MOV10, UPF1, and HuR. (D) Co-IP experiments were performed on HEK293 cells after overexpression of GFP-IRAV (IRAV) or GFP-CAT (Control). Cell lysates were either left untreated (−) or treated with RNase A (+), followed by IP with an antibody for GFP. Western blots were then performed for IRAV putative interaction partners MOV10, UPF1, and HuR.
To further explore interactions between IRAV and proteins associated with RNA processing, MS hits were ranked based on the spectral count, and the top-scoring proteins were further characterized (see Table S1 in the supplemental material). We were able to verify interactions between IRAV and MOV10, UPF1, and HuR by co-IP of overexpressed IRAV, followed by Western blotting (Fig. 4C). Because MOV10, UPF1, and HuR are all known to be RNA binding proteins, we examined the influence of RNase A treatment on these interactions. RNase treatment resulted in either complete or partial loss of interactions with IRAV (Fig. 4D). Partial loss of interactions due to RNase A treatment was previously described for interactions between MOV10 and UPF1 (36).
IRAV colocalizes with P bodies.
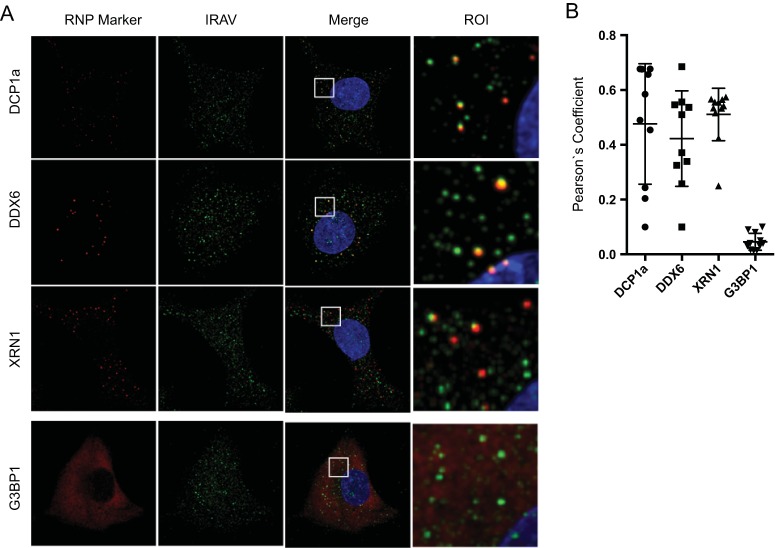
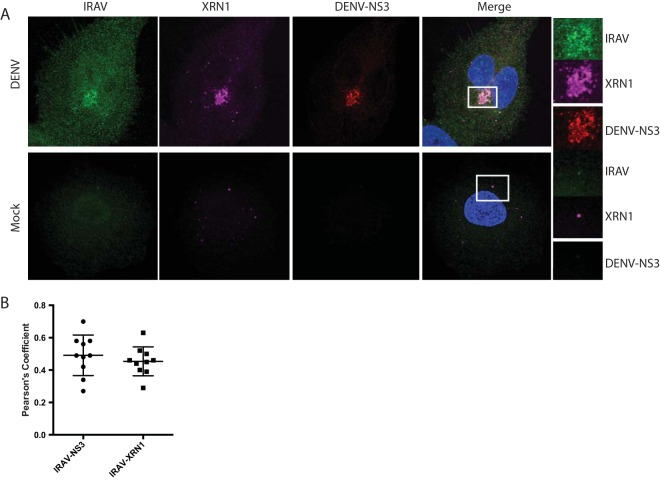
Because many of the RNA binding proteins shown to interact with IRAV are associated with ribonucleoprotein (RNP) granules, we next investigated whether IRAV colocalized with specific RNP granule markers. Here, we examined A549 cells for the localization of P body markers (XRN1, DCP1a, and DDX6/RCK), as well as the stress granule marker G3BP1 (Fig. 5A). We observed colocalization between IRAV and all three P body markers in IFN-treated cells; however, no statistically significant colocalization was observed between IRAV and G3BP1, indicating that IRAV associated with P bodies in the cytoplasm of IFN-treated cells (Fig. 5B). To characterize the association of IRAV with P body components after DENV infection, A549 cells were infected with DENV, followed by staining for IRAV and XRN1 (Fig. 6A and B). As previously described for XRN1 (37), IRAV relocalized to the DENV replication complex, colocalizing with DENV NS3.
FIG 5.
IRAV associates with P bodies in IFN-β-treated cells. (A) Confocal microscopy of IRAV; P body markers DCP1a, DDX6, and XRN1; and the stress granule marker G3BP1a in A549 cells after treatment with IFN-β (10 ng/ml) for 16 h. Green, IRAV; red, RNP markers. The nucleus was stained with DAPI (blue). Regions of interest (ROI) are boxed in white. (B) Colocalization coefficients of IRAV with DCP1a, DDX6, XRN1, and G3BP1 as determined by Pearson's linear correlation coefficient. The error bars represent standard deviations.
FIG 6.
IRAV relocalizes to the replication complex after DENV infection. (A) Confocal microscopy of XRN1 colocalized with IRAV and DENV NS3 at the replication complex in DENV-infected or mock-infected A549 cells. Green, IRAV; magenta, XRN1; red, DENV NS3. The nucleus was stained with DAPI (blue). Colocalization between IRAV, XRN1, and DENV NS3 is shown in white. Regions of interest (ROI) are boxed in white. (B) Colocalization coefficients of IRAV and DENV NS3 or IRAV and XRN1 in DENV-infected A549 cells as determined by Pearson's linear correlation coefficient, demonstrating colocalization between IRAV and both XRN1 and DENV NS3 in DENV-infected cells. The error bars represent standard deviations.
IRAV interacts with MOV10.
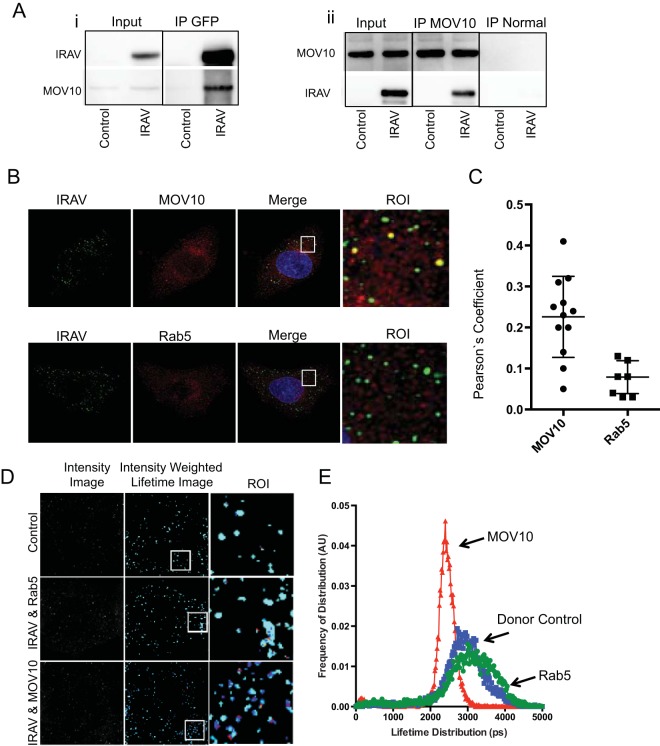
Due to its known role as a restriction factor for retroviruses (38–41), hepatitis C virus (6), and hepatitis B virus (42), we further characterized interactions between MOV10 and IRAV. Co-IPs and reciprocal co-IPs were performed using HEK293 cells transfected with either IRAV (GFP-IRAV) or a negative control (GFP-CAT). Co-IPs were then performed using an antibody either to overexpressed IRAV or to endogenous MOV10, and Western blots were analyzed for the presence of either MOV10 or IRAV (Fig. 7A). In addition, confocal microscopy and fluorescence resonance energy transfer (FRET) by fluorescence lifetime imaging (FLIM) analyses were used to characterize interactions between endogenous IRAV and MOV10 in A549 cells. Confocal microscopy revealed colocalization between IRAV and MOV10, as determined by Pearson's linear correlation coefficient (Fig. 7B and C). Moreover, FRET-by-FLIM analysis demonstrated interactions between IRAV and MOV10, with lifetime values of 1.8 to 2.1 compared to values of 2.5 to 2.8 for the donor control, representing 25% to 28% FRET efficiency. The endosomal marker Rab5 was used as a negative control. Rab5 did not demonstrate significant colocalization with IRAV or a shift in lifetime values (Fig. 7D and E).
FIG 7.
IRAV interacts with MOV10. (A) Co-IP experiments performed on HEK293 cells transfected with expression vectors for GFP-IRAV (IRAV) or GFP-CAT (Control), followed by IP with either an antibody for GFP (i) or endogenous MOV10 (ii). Normal IgG was used as a negative control for endogenous MOV10. Western blots were then performed for IRAV or MOV10. (B) Confocal microscopy of IRAV and MOV10 or Rab5 in IFN-β-treated A549 cells. Green, IRAV; red, MOV10 or Rab5. The nucleus was stained with DAPI (blue). ROI are boxed in white. (C) Colocalization coefficients of IRAV and MOV10 or Rab5 in IFN-β-treated A549 cells as determined by Pearson's linear correlation coefficient. (D) Representative image of FRET-by-FLIM analysis of IRAV interactions with MOV10 or Rab5 in IFN-β-treated A549 cells, showing the image intensity, intensity-weighted lifetime, and selected ROI. (E) Lifetime distribution plot versus frequency of distribution, in arbitrary units (AU), of FRET-by-FLIM analysis of IRAV (donor control), IRAV and MOV10, or IRAV and Rab5 in IFN-β-treated A549 cells. The plot was constructed using data from 10 images each, collected from two independent experiments. IRAV interactions with MOV10 are shown in red, Rab5 in green, and the donor control in blue. The error bars represent standard deviations.
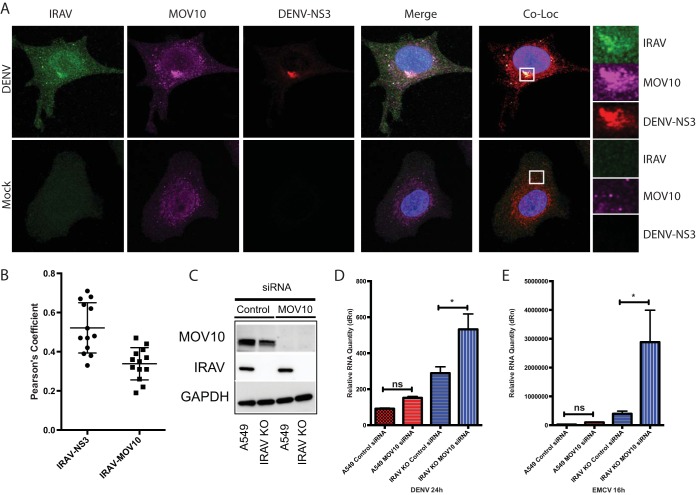
Confocal microscopy was also used to examine localization of IRAV and MOV10 in DENV-infected and mock-infected A549 cells (Fig. 8A and B). Here, it was observed that, similar to XRN1, both IRAV and MOV10 relocalized to the viral replication complex, colocalizing with DENV NS3. To further substantiate a role for MOV10 in restriction of DENV replication, siRNA-mediated knockdown experiments were performed in A549 and IRAV KO cells (Fig. 8C). Depletion of MOV10 resulted in an increase in virus replication for DENV, as well as for EMCV, which was significantly greater in IRAV KO cells than in IRAV-expressing A549 cells (Fig. 8D and E), indicating that both IRAV and MOV10 play roles in the restriction of virus replication.
FIG 8.
MOV10 localizes to the DENV replication complex and restricts viral replication. (A) Confocal microscopy of MOV10 colocalized with IRAV and DENV NS3 at the replication complex in DENV-infected or mock-infected A549 cells. Green, IRAV; magenta, MOV10; red, DENV NS3. The nucleus was stained with DAPI (blue). Colocalization between IRAV, MOV10, and DENV NS3 is shown in white. ROI are boxed in white. (B) Colocalization coefficients of IRAV and DENV NS3 or IRAV and MOV10 in DENV-infected A549 cells as determined by Pearson's linear correlation coefficient, demonstrating colocalization between IRAV and both MOV10 and DENV NS3. (C) Western blot analysis of siRNA-mediated knockdown of MOV10 in A549 or IRAV KO cells. The cells were treated with a gene-specific siRNA for MOV10 (Qiagen Hs_MOV10_5 FlexiTube siRNA) or a negative-control siRNA for 48 h, followed by Western blot analysis for MOV10 or IRAV. GAPDH was used as a loading control. (D) qRT-PCR analysis of A549 or IRAV KO cells treated with either negative-control siRNA or gene-specific MOV10 siRNA for 48 h. The cells were then infected with DENV (MOI, 0.1) for 24 h and analyzed for DENV RNA. Samples were normalized to the HPRT housekeeping gene, and the change in expression was calculated relative to time zero. (E) qRT-PCR analysis of A549 or IRAV KO cells treated with either negative-control siRNA or gene-specific MOV10 siRNA for 48 h. The cells were then infected with EMCV (MOI, 0.001) for 24 h and analyzed for EMCV RNA. Samples were normalized to the HPRT housekeeping gene, and the change in expression was calculated relative to time zero. The error bars represent standard deviations. *, P < 0.05; ns, not significant.
DISCUSSION
Interferon-stimulated genes are key mediators of antiviral immunity and play a pivotal role in the immune response to DENV infection. While the IFN response is an innate and broad-spectrum response to many pathogens, ISGs have been demonstrated to have remarkable specificity both for the viruses they target and the pathways through which they inhibit viral replication (6). Hundreds of ISGs have been shown to be upregulated in response to induction of IFN signaling pathways, illustrating the scope and complexity of the IFN response. To date, only a fraction of these genes have been characterized.
Here, we show that IRAV is an IFN-stimulated gene with antiviral activity against DENV. Knockout of the IFN signaling component IRF9 results in a significant reduction of IRAV expression after IFN treatment, suggesting regulation through the canonical ISGF3 complex; however, it should be noted that there appears to be some baseline constitutive expression of IRAV. In addition, IRAV homologues were identified in hemichordates and echinoderms, organisms that lack a “classical” IFN response, suggesting the possibility of IFN-independent functions for IRAV-like proteins.
IRAV associates with P bodies, as evidenced by interactions with P body-associated proteins (MOV10 and UPF1) and colocalization with P body markers (DCP1a, XRN1, and DDX6). P bodies are discrete cytoplasmic structures composed of RNA and proteins that are associated with various aspects of RNA turnover, including nonsense-mediated decay, adenylate-uridylate-rich element (ARE)-mediated decay, and gene silencing (43). Notably, both DENV and West Nile virus (WNV) have been shown to associate with components of P bodies. Subgenomic flavivirus RNA (sfRNA) inhibits the activity of XRN1 and TRIM25 (44–46), and DDX6 interacts with DENV untranslated regions (UTRs) and localizes to DENV replication complexes, possibly playing a role in virus replication (47). Furthermore, WNV infection of HeLa cells results in a decrease of both the size and number of P bodies, along with a relocalization of P body components (including GW182, DDX3, and XRN1) to WNV replication complexes, colocalizing with NS3 (37). This is similar to what we observed with IRAV during DENV infection, with IRAV puncta becoming more diffuse and colocalizing with NS3 in distinct perinuclear complexes. A number of other viruses, including hepatitis C virus (48), poliovirus (49), influenza virus (50), and bunyaviruses (51, 52), have been shown to interact with P bodies or with P body components (53).
MOV10 and UPF1 are both members of the SF1 family of helicases and have been previously shown to interact with one another and to form complexes with APOBEC3G and Argonaute 2 (40, 54), as well as with the antiviral protein ZAP (55). Interactions between MOV10 and UPF1 are partially sensitive to RNase treatment (36), as we have also demonstrated for IRAV interactions with MOV10 or UPF1. Knockdown of MOV10 results in an increase in the half-lives of mRNA targets (36). MOV10 has been shown to be involved in retrotransposition of LINE elements (56–58), as well as in microRNA pathways (54, 59). MOV10 may also play a role in mediating antiviral response (60) and restricts replication of retroviruses (38–41), hepatitis C virus (6), hepatitis B virus (42), and influenza virus (61). UPF1 is a key component of nonsense-mediated RNA decay pathways (62) and has been demonstrated to play a role in viral replication cycles (63, 64). IRAV also associates with the RNA binding protein HuR. HuR has been shown to be involved in RNA decay pathways (65) and has been linked to viral replication and modulation of the host response (66–69). In addition, IRAV has been previously shown to interact with RNA binding proteins LARP1 and PABPC1, both of which have also been shown to be involved in RNA decay (23). Taken together, IRAV's role as an RNA binding protein, its association with proteins linked to RNA decay, and its localization to P bodies (sites of RNA decay) and the marked increase in viral RNA observed after knockout of IRAV all suggest a role for IRAV in degradation of viral RNA. The role of MOV10 in IRAV-mediated antiviral activity remains unclear; however, given MOV10's role as an RNA helicase involved in RNA decay pathways, as well as its localization at the viral replication complex and the increase in viral RNA observed after its knockdown, MOV10 may function in conjunction with IRAV and other proteins to aid in destabilization of viral RNA.
In conclusion, we show that IRAV is an ISG that is regulated through the canonical type I interferon signaling pathway. IRAV displays antiviral activity against DENV and EMCV and interacts with DENV proteins. IRAV is an RNA binding protein that localizes to P bodies, sites of RNA decay. Additionally, IRAV interacts with MOV10 and UPF1, two proteins previously shown to interact with each other and to be involved in RNA decay pathways. These data serve to identify an ISG with antiviral activity against DENV, as well as to suggest a mechanism of action involving the destabilization of viral RNA.
MATERIALS AND METHODS
Viruses and cell culture.
The DENV serotype 2 isolate Tonga/74 was generously provided by S. Whitehead (Laboratory of Infectious Diseases [LID], NIAID) and passaged in C6/36 cells. The titer of virus was determined on Vero cells as previously described (70, 71). EMCV and vesicular stomatitis virus (VSV) were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) and passaged in Vero cells. Virus infections were performed at the indicated MOI, and virus was allowed to adsorb to the cells for 1 hour. Unbound virus was then removed, and the cells were washed with phosphate-buffered saline (PBS), followed by the addition of RPMI medium containing 10% fetal bovine serum. Primary human monocytes were obtained using the Gambro Elutra method from the NIH Clinical Research Center Department of Transfusion. Monocyte-derived macrophages were cultured as previously described (72). HEK293 cells were obtained from Life Technologies (Grand Island, NY). Daudi cells (a human B lymphoblast line) were obtained from P. Grimley (Department of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD). The human epithelial breast adenocarcinoma cell line MDA-MB-231 was obtained from Raj Puri (FDA, Center for Biologics Evaluation and Research [CBER], Bethesda, MD), OVCAR3 (human ovarian adenocarcinoma) cells were obtained from the National Cancer Institute (Bethesda, MD). Human cervical carcinoma HeLa S3 cells (CCL-2.2); a human monocyte-like cell line derived from histiocytic lymphoma (U937 [CRL-1593.2]); and Detroit 551 (CCL-110), NCI-H1299 (CRL-5803), HEL 299 (CCL-137), and human lung carcinoma A549 (CCL-185) cell lines were obtained from the ATCC. All the cell lines were cultured in RPMI medium containing 10% fetal bovine serum and 50 μg/ml of penicillin and streptomycin. IFN-β1a (Avonex), with a specific activity of 200 million IU per mg, was obtained from Biogen Idec (Cambridge, MA).
Antibodies and fluorescent labeling.
Primary antibodies were obtained as follows: C19orf66 (FLJ11286; HPA042001), Sigma-Aldrich Corp., St. Louis, MO; UPF1 (D15G6), Cell Signaling Technology, Inc., Danvers, MA; Rab5 (C8B1), Cell Signaling Technology, Inc., Danvers, MA; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (D16H11), Cell Signaling Technology, Inc., Danvers, MA; MOV10 (A301-571A), Bethyl Laboratories, Inc., Montgomery, TX; XRN1 (C-1), Santa Cruz Biotechnology, Inc., Dallas, TX; HuR (G-8), Santa Cruz Biotechnology, Inc., Dallas, TX; hDcp1a (65-Y), Santa Cruz Biotechnology, Inc., Dallas, TX; RCK (DDX6; E-12), Santa Cruz Biotechnology, Inc., Dallas, TX; G3BP1 (H-10), Santa Cruz Biotechnology, Inc., Dallas, TX; IRF9 (ISGF3γ; 610285), BD Biosciences, San Jose, CA; IFIT3, Covance, Inc., Princeton, NJ; NS4A (SAB2700179), Sigma-Aldrich Corp., St. Louis, MO; NS3 (SAB2700181), Sigma-Aldrich Corp., St. Louis, MO; and E (SAB2700196), Sigma-Aldrich Corp., St. Louis, MO.
Horseradish peroxidase (HRP)-labeled secondary antibodies for Western blotting were obtained from Santa Cruz Biotechnology, Inc. Fluorescently labeled secondary antibodies for confocal microscopy were obtained from Life Technologies (Grand Island, NY). Fluorescent labeling of primary antibodies for colocalization and FRET-by-FLIM of IRAV and MOV10, Rab5, DENV NS3, and DENV NS4A were performed using Zenon antibody-labeling kits (Life Technologies) according to the manufacturer's specifications.
Plasmid constructs and transfections.
A plasmid containing the IRAV gene was purchased from GeneCopeia (Rockville, MD) and amplified using directional forward and reverse primers (5′-CAC CAT GTC TCA GGA AGG TGT GGA-3′ and 5′-GGC GGG CCC AGG GAG TGA-3′), followed by cloning into a pENTR/D-TOPO vector (Life Technologies, Grand Island, NY). The IRAV gene sequence was then transferred to a pcDNA 6.2/N-EmGFP-DEST vector (Life Technologies) via LR recombination using Gateway LR Clonase enzyme mix (Life Technologies) according to the manufacturer's protocol. The constructs were sequenced to verify correct sequence and orientation. Negative-control vectors were obtained from Life Technologies. Transfections were performed in HEK293 cells using Opti-MEM and X-tremeGene HP according to the manufacturer's directions (Roche Diagnostics, Indianapolis, IN).
CRISPR/Cas9 knockout and siRNA knockdown experiments.
CRISPR/Cas9 knockout cell lines were generated using A549 cells. An FLJ11286/C19orf66 (IRAV) CRISPR/Cas9 knockout plasmid (sc-408037), containing the Cas9 nuclease and guide RNA, and its complementary homology-directed repair plasmid (sc-408037), containing red fluorescent protein and a puromycin resistance gene flanked by 5′ and 3′ arms homologous to regions flanking the Cas9 target site, or an IRF9 CRISPR/Cas9 knockout plasmid (sc-400958) and a homology-directed repair plasmid (sc-400958), were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX). A549 cells were cotransfected with CRISPR/Cas9 and homology-directed repair plasmids using GenMute transfection reagent (SignaGen Laboratories, Ijamsville, MD), and knockouts were screened for puromycin resistance (3 μg/μl). Individual subclones were then assayed for gene expression by both PCR and Western blotting. Knockdown experiments were performed using gene-specific siRNAs. IRAV siRNA (Hs_FLJ11286_5; catalog number S104203668), MOV10 siRNA (Hs_MOV10_5 FlexiTube siRNA), and AllStars negative-control siRNA (SI03650318) were obtained from Qiagen (Valencia, CA). Reverse transfections were performed using Lipofectamine RNAiMax transfection reagent (Life Technologies), according to the manufacturer's specifications.
Immunoprecipitation, mass spectrometry, and Western blot analysis.
Protein coimmunoprecipitations of GFP-IRAV- or GFP-CAT-transfected cells and Western blot analyses were performed as previously described (73). Identification of gel-separated proteins was performed on reduced and alkylated, trypsin-digested samples. The supernatant and two washes (5% formic acid in 50% acetonitrile) of the gel digests were pooled and concentrated by speed vac (Labconco, Kansas, MO) to dryness directly in 200-μl polypropylene autosampler vials (Sun Sri, Rockwood, TN). The recovered peptides were resuspended in 5 μl of solvent A (0.1% formic acid, 2% acetonitrile, and 97.9% water). Prior to mass spectrometry analysis, the resuspended peptides were chromatographed directly on column, without trap cleanup. The bound peptides were separated at 500 nl/min, generating 80 × 105 to 120 × 105 Pa pressure, using an AQ C18 reverse-phase medium (3-μm particle size and 200-μm pore size) packed in a pulled-tip nanochromatography column (0.100-mm inside diameter [i.d.] by 150-mm length) from Precision Capillary Columns (San Clemente, CA). The chromatography was performed in line with an LTQ-Velos Orbitrap mass spectrometer (ThermoFisher Scientific, West Palm Beach, FL), and the mobile phase consisted of a linear gradient prepared from solvent A and solvent B (0.1% formic acid, 2% water, and 97.9% acetonitrile) at room temperature. Nano-liquid chromatography (LC)-MS (LC–tandem MS [MS-MS]) was performed with a ProXeon Easy-nLC II multidimensional liquid chromatograph and a temperature-controlled Ion Max Nanospray source (ThermoFisher Scientific) in line with the LTQ-Velos Orbitrap mass spectrometer.
A Mascot (Matrix Science, Beachwood, OH) search of the data was performed against a concatenated sequence file containing dengue virus proteins found in the UniProtKB TrEMBL database, human protein UniProtKB Swiss-Prot, and the common contaminant proteins found in the GPM.org's cRAP database, all downloaded in March 2014. The data were searched with 2 allowed missed cleavages and mass tolerances of 15 ppm and 0.8 Da for the precursor and fragment ions, respectively. Carbamidomethylation of cysteine was set as a fixed modification, while oxidation of methionine and deamidation of asparagine and glutamine were searched as dynamic modifications. The resulting search files were reclustered against the same sequence database for further analysis using ProteoIQ software (Premier Biosoft, Palo Alto, CA). Filters of 2 spectra per peptide sequence and 2 peptides per protein were in place for protein assignments. The data were then ranked based on the number of peptide hits and the spectral count.
Quantitative real-time PCRs.
RNA was extracted from DENV-infected cells using a Qiagen RNeasy minikit and QIAshredder spin columns and including DNase digestion (Qiagen, Valencia, CA). qRT-PCRs were performed using the SensiFast Probe No-ROX One-Step kit (Bioline USA Inc., Taunton, MA), and reactions were run on a Stratagene Mx3005P real-time thermocycler (Agilent Technologies, Santa Clara, CA). Primers and probes were obtained from Integrated DNA Technologies, Inc. (Coralville, IA), and were as follows: DENV-2 (forward, 5′-TGC CTA CAG TTC TAC GTC TCC-3′; reverse, 5′-TCG TTT CCT AAC AAT CCC ACC-3′; probe, 5′-6-carboxyfluorescein (FAM)-CCT TCC AAT-ZEN-CTC TTT CCT GAA GCC TCT C-Iowa black fluorescence quencher [IABkFQ]-3′), EMCV (forward, 5′-TTC AGC GTT TTC TAC TCC CTG-3′; reverse, 5′-TCA CTC CCC TCA CTT ACC C-3′; probe, 5′-FAM-AGA AAT CCT-ZEN-TCC CTG CGC TCA CC-IABkFQ-3′), IFN-β (forward, 5′-TGA AGC AAT TGT CCA GTC CC-3′; reverse, 5′-GCC AAG GAG TAC AGT CAC TG-3′; probe, 5′-FAM-AGG CAC AGG-ZEN-CTA GGA GAT CTT CAG T-IABkFQ-3′), IRAV (forward, 5′-CTA AGT AAC GAT CTG GAT GCC C-3′; reverse, 5′-CGT TGA AAC ATG CGT AGG TTG-3′; probe, 5′-hexachlorofluorescein [HEX]-CCT GAA TGT-ZEN-CCC GGT CCT GCT T-IABkFQ-3′), HPRT (forward, 5′-GTA TTC ATT ATA GTC AAG GGC ATA TCC-3′; reverse, 5′-AGA TGG TCA AGG TCG CAA G-3′; probe, 5′-Cy5-TGG TGA AAA GGA CCC CAC GAA GT-Iowa black RQ-Sp [IAbRQSp]-3′), IRF9 (forward, 5′-CAA AGG CCT GCT CCA TCT-3′; reverse, 5′-TGC AGA GAC TTG GTC AGG TA-3′; probe, 5′-FAM-CAT GGC TCT-ZEN-CTT CCC AGA AAT TCA GTG T-IABkFQ-3′), MX1 (forward, 5′-CCA CCC ATA TTT CAG GGA TCT G-3′; reverse, 5′-TCT GGT GAG TCT CCT TGA TTT G-3′; probe, 5′-HEX-TGT GTG ATG AGC TCG CTG GTA AGT TT-IABkFQ-3′), and IFIT3 (forward, 5′-CAC TGT CTT CCT TGA ATA AGT TCC-3′; reverse, 5′-AGA ACA AAT CAG CCT GGT CAC-3′; probe, 5′-HEX-AGA AAA TCC-ZEN-TTC CAC AGC TGA AAT GCC-IABkFQ-3′).
Protein expression, purification, and fluorescence polarization.
The nucleotide sequence representing the first 269 aa of IRAV was amplified by PCR using forward and reverse primers (5′-CTA GTC GAC ATG TCT CAG GAA GGT GTG GAG CTG GAG-3′ and 5′-CTA GCG GCC GCC AGG ATG AGG TTG TCC AGG TCT TCC AGG AGG-3′) and ligated into a modified MBP-tagged expression vector with a pET30a backbone. The BL-21-CodonPlus RIPL strain (Stratagene, Santa Clara, CA) was then transformed with the plasmid, and recombinant protein was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 18°C for 4 h. The soluble fraction of the recombinant protein was purified by Ni-nitrilotriacetic acid (NTA) chromatography, followed by Superdex-200 gel filtration with 200 mM NaCl, 20 mM HEPES, pH 7.4. The nonaggregate fraction of the His-MBP-tagged IRAV protein was analyzed by SDS-PAGE as ∼95% pure.
For FP assays, the following FAM-labeled 30-mer RNA probes were used: 5′-rArGrU rUrGrU rUrArG rUrCrU rArCrG rUrGrG rArC/36-FAM-3′, with or without the reverse complement 5′-rGrUrC rCrArC rGrUrA rGrArC rUrArA rCrArA rCrU-3′ (where “r” refers to ribose); or DNA probes 5′-AGT TGT TAG TCT ACG TGG AC-FAM-3′, with or without the reverse complement 5′-GTC CAC GTA GAC TAA CAA CT-3′.
Oligomers were dissolved in binding buffer (100 mM KCl, 20 mM HEPES, pH 7.4), heated to 85°C for 10 min, and allowed to cool slowly. The oligomers were then added to serial dilutions of protein solution in triplicate, and FP was measured on a Corning black 96-well, half-area assay plate (Corning, NY) using a Hidex Sense Microplate Reader (Turku, Finland).
Confocal microscopy and fluorescence resonance energy transfer by fluorescence lifetime imaging.
A549 cells were plated on poly-d-lysine-coated 35-mm culture dishes (MatTek, Ashland, MA) and either treated with IFN-β (10 ng/ml) for 16 h or infected with DENV-2 (MOI, 3) for 48 h. The cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.1% or 0.5% Triton X-100, followed by blocking with 5% bovine serum albumin (BSA) for 30 min. Primary antibodies were diluted in 10% normal goat serum and incubated at room temperature for 2 h, followed by three washes in PBS and staining with fluorescently labeled secondary antibody (1:500) and nuclear DAPI (4′,6-diamidino-2-phenylindole) stain (Life Technologies). Primary labeled antibodies were diluted in 10% normal goat serum and incubated at room temperature for 2 h, followed by three washes in PBS, and postfixed with 4% paraformaldehyde in PBS. Images were collected on a Leica SP8 inverted confocal microscope with a 63× oil immersion objective (Leica Microsystems, Buffalo Grove, IL). Colocalization analysis was performed using Imaris software (Bitplane Inc., South Windsor, CT). FRET-by-FLIM analysis was performed as previously described (73).
Gene ontology and statistical analyses.
GO analysis was performed using proteins identified by MS with spectral counts above 50. Selected protein accessions were analyzed using the PANTHER classification system (http://pantherdb.org) (35). All statistical analyses were performed on Prism (GraphPad Software, Inc.), using one-way analysis of variance (ANOVA) with Tukey's post hoc test and a P value of 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tsan Xiao and David Garboczi for their insights into protein structure; Ming Zhao and Renee Olano from the Research Technologies Branch, NIAID, NIH, for excellent technical assistance; Yajuan Li from the School of Life Sciences, University of Science and Technology of China, for assistance with protein purification; Daniel Green for providing human monocytes; Xavier Ambroggio and Vijayaraj Nagarajan for their help with computational analysis; Emerito Amaro-Carambot for technical expertise; Stephen Whitehead for providing reagents and support; and Sonja Best and Ted Pierson for their valuable insight and critical readings of the manuscript.
This work was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01606-16.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat Rev Microbiol 8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyle JL, Harris E. 2008. Global spread and persistence of dengue. Annu Rev Microbiol 62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 4.WHO. 2006. WHO scientific working group report on dengue; reference number TDR/SWG/08. WHO, Geneva, Switzerland. [Google Scholar]
- 5.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoggins JW. 2014. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol 6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warke RV, Martin KJ, Giaya K, Shaw SK, Rothman AL, Bosch I. 2008. TRAIL is a novel antiviral protein against dengue virus. J Virol 82:555–564. doi: 10.1128/JVI.01694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brass AL, Huang I-C, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, Beard MR. 2013. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl Trop Dis 7:e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang D, Weidner JM, Qing M, Pan X-B, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi P-Y, Block TM, Guo J-T. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Li NL, Wang J, Shi P-Y, Wang T, Miller MA, Li K. 2014. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J Virol 88:13821–13835. doi: 10.1128/JVI.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho H, Shrestha B, Sen GC, Diamond MS. 2013. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol 87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. 2011. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol 85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Sun J, Meng L, Heathcote J, Edwards AM, McGilvray ID. 2010. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J Gen Virol 91:382–388. doi: 10.1099/vir.0.015388-0. [DOI] [PubMed] [Google Scholar]
- 16.Sadeghi F, Bokharaei-Salim F, Salehi-Vaziri M, Monavari SH, Alavian SM, Salimi S, Vahabpour R, Keyvani H. 2014. Associations between human TRIM22 gene expression and the response to combination therapy with Peg-IFNα-2a and ribavirin in Iranian patients with chronic hepatitis C. J Med Virol 86:1499–1506. doi: 10.1002/jmv.23985. [DOI] [PubMed] [Google Scholar]
- 17.Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. 2011. TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10:185–196. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P-Y, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Ding SC, Cho H, Chung BC, Gale M, Chanda SK, Diamond MS. 2013. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. mBio 4:e00385-00313. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, Zoon KC. 2010. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J Virol 84:10671–10680. doi: 10.1128/JVI.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balinsky CA, Schmeisser H, Zoon KC. 2014. Characterization of a novel interferon stimulated gene with antiviral activity. Cytokine 70:28–29. [Google Scholar]
- 22.Balinsky CA, Schmeisser H, Ganesan S, Jin T, Singh K, Zoon KC. 2015. Characterization of a novel interferon stimulated gene with antiviral activity against dengue virus. Cytokine 76:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki Y, Chin W-X, Han Q, Ichiyama K, Lee CH, Eyo ZW, Ebina H, Takahashi H, Takahashi C, Tan BH, Hishiki T, Ohba K, Matsuyama T, Koyanagi Y, Tan Y-J, Sawasaki T, Chu JJH, Vasudevan SG, Sano K, Yamamoto N. 2016. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog 12:e1005357. doi: 10.1371/journal.ppat.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh MK, Scott TF, LaFramboise WA, Hu FZ, Post JC, Ehrlich GD. 2007. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J Neurol Sci 258:52–59. doi: 10.1016/j.jns.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Taylor MW, Tsukahara T, McClintick JN, Edenberg HJ, Kwo P. 2008. Cyclic changes in gene expression induced by Peg-interferon alfa-2b plus ribavirin in peripheral blood monocytes (PBMC) of hepatitis C patients during the first 10 weeks of treatment. J Transl Med 6:66. doi: 10.1186/1479-5876-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmerer JM, Lesinski GB, Ruppert AS, Radmacher MD, Noble C, Kendra K, Walker MJ, Carson WE. 2008. Gene expression profiling reveals similarities between the in vitro and in vivo responses of immune effector cells to interferon-alpha. Clin Cancer Res 14:5900–5906. doi: 10.1158/1078-0432.CCR-08-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD, Balderas RS, Wilkinson P, Pantaleo G, Tartaglia J, Haddad EK, Sékaly R-P. 2008. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey SAK, Romanowski EG, Yates KA, Gordon YJ. 2005. Adenovirus-directed ocular innate immunity: the role of conjunctival defensin-like chemokines (IP-10, I-TAC) and phagocytic human defensin-alpha. Invest Ophthalmol Vis Sci 46:3657–3665. doi: 10.1167/iovs.05-0438. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, Mason RJ. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am J Respir Cell Mol Biol 45:582–591. doi: 10.1165/rcmb.2010-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zapata JC, Carrion R, Patterson JL, Crasta O, Zhang Y, Mani S, Jett M, Poonia B, Djavani M, White DM, Lukashevich IS, Salvato MS. 2013. Transcriptome analysis of human peripheral blood mononuclear cells exposed to Lassa virus and to the attenuated Mopeia/Lassa reassortant 29 (ML29), a vaccine candidate. PLoS Negl Trop Dis 7:e2406. doi: 10.1371/journal.pntd.0002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kash JC, Mühlberger E, Carter V, Grosch M, Perwitasari O, Proll SC, Thomas MJ, Weber F, Klenk H-D, Katze MG. 2006. Global suppression of the host antiviral response by Ebola- and Marburg viruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol 80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bull TM, Meadows CA, Coldren CD, Moore M, Sotto-Santiago SM, Nana-Sinkam SP, Campbell TB, Geraci MW. 2008. Human herpesvirus-8 infection of primary pulmonary microvascular endothelial cells. Am J Respir Cell Mol Biol 39:706–716. doi: 10.1165/rcmb.2007-0368OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazaki D, Haruki T, Takeda S, Sasaki S-I, Yakura K, Terasaka Y, Komatsu N, Yamagami S, Touge H, Touge C, Inoue Y. 2011. Herpes simplex virus type 1-induced transcriptional networks of corneal endothelial cells indicate antigen presentation function. Invest Ophthalmol Vis Sci 52:4282–4293. doi: 10.1167/iovs.10-6911. [DOI] [PubMed] [Google Scholar]
- 34.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, André P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol Syst Biol 4:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Chahar HS, Chen S, Manjunath N. 2013. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology 436:1–7. doi: 10.1016/j.virol.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu W-S, Pathak VK. 2010. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J Virol 84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, Kewalramani VN, Unutmaz D. 2010. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS One 5:e9081. doi: 10.1371/journal.pone.0009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izumi T, Burdick R, Shigemi M, Plisov S, Hu W-S, Pathak VK. 2013. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol 87:11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, Zheng Y-H. 2010. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem 285:14346–14355. doi: 10.1074/jbc.M110.109314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y- X, Li D, Fu L-J, Fu B-Q, Chen S-J, Xu W-Z, Teng X, Song Z-W, Gu H-X. 2015. The role of Moloney leukemia virus 10 in hepatitis B virus expression in hepatoma cells. Virus Res 197:85–91. doi: 10.1016/j.virusres.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife 3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. 2012. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol 8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Vilaró G, Fernández-Carrillo C, Mensa L, Miquel R, Sanjuan X, Forns X, Pérez-Del-Pulgar S, Díez J. 2015. Hepatitis C virus infection inhibits P-body granule formation in human livers. J Hepatol 62:785–790. doi: 10.1016/j.jhep.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Dougherty JD, White JP, Lloyd RE. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol 85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mok BW-Y, Song W, Wang P, Tai H, Chen Y, Zheng M, Wen X, Lau S-Y, Wu WL, Matsumoto K, Yuen K-Y, Chen H. 2012. The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA-associated protein 55 (RAP55) during virus infection. J Virol 86:12695–12707. doi: 10.1128/JVI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng E, Mir MA. 2012. Signatures of host mRNA 5′ terminus for efficient hantavirus cap snatching. J Virol 86:10173–10185. doi: 10.1128/JVI.05560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hopkins K, Cherry S. 2013. Bunyaviral cap-snatching vs. decapping: recycling cell cycle mRNAs. Cell Cycle 12:3711–3712. doi: 10.4161/cc.26878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd RE. 2013. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip Rev RNA 4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Zhang X, Huang F, Yang B, Li J, Liu B, Luo H, Zhang P, Zhang H. 2012. APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J Biol Chem 287:29373–29383. doi: 10.1074/jbc.M112.354001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH. 2015. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 11:e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goodier JL, Cheung LE, Kazazian HH. 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, Guo F, Liang C, Cen S. 2013. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem 288:21148–21160. doi: 10.1074/jbc.M113.465856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor MS, Lacava J, Mita P, Molloy KR, Huang CRL, Li D, Adney EM, Jiang H, Burns KH, Chait BT, Rout MP, Boeke JD, Dai L. 2013. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155:1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S. 2014. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep 9:1729–1741. doi: 10.1016/j.celrep.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cuevas RA, Ghosh A, Wallerath C, Hornung V, Coyne CB, Sarkar SN. 2016. MOV10 provides antiviral activity against RNA viruses by enhancing RIG-I-MAVS-independent IFN induction. J Immunol 196:3877–3886. doi: 10.4049/jimmunol.1501359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Huang F, Tan L, Bai C, Chen B, Liu J, Liang J, Liu C, Zhang S, Lu G, Chen Y, Zhang H. 2016. Host protein Moloney leukemia virus 10 (MOV10) acts as a restriction factor of influenza A virus by inhibiting the nuclear import of the viral nucleoprotein. J Virol 90:3966–3980. doi: 10.1128/JVI.03137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imamachi N, Tani H, Akimitsu N. 2012. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug Discov Ther 6:55–61. [PubMed] [Google Scholar]
- 63.Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, Mühlemann O, Azzalin C, Helenius A. 2014. The host nonsense-mediated mRNA decay pathway restricts mammalian RNA virus replication. Cell Host Microbe 16:403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 64.Serquiña AKP, Das SR, Popova E, Ojelabi OA, Roy CK, Göttlinger HG. 2013. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol 87:8853–8861. doi: 10.1128/JVI.00925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Ding N, Guo J, Xia J, Ruan Y. 2016. Dysregulation of TTP and HuR plays an important role in cancers. Tumour Biol 37:14451–14461. doi: 10.1007/s13277-016-5397-z. [DOI] [PubMed] [Google Scholar]
- 66.Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. 2013. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep 5:909–917. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. 2015. HuR displaces polypyrimidine tract binding protein to facilitate La binding to the 3′ untranslated region and enhances hepatitis C virus replication. J Virol 89:11356–11371. doi: 10.1128/JVI.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hung C-M, Huang W-C, Pan H-L, Chien P-H, Lin C-W, Chen L-C, Chien Y-F, Lin C-C, Leow K-H, Chen W-S, Chen J-Y, Ho C-Y, Hou P-S, Chen Y-J. 2014. Hepatitis B virus X upregulates HuR protein level to stabilize HER2 expression in hepatocellular carcinoma cells. Biomed Res Int 2014:827415. doi: 10.1155/2014/827415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin J-Y, Brewer G, Li M-L. 2015. HuR and Ago2 bind the internal ribosome entry site of enterovirus 71 and promote virus translation and replication. PLoS One 10:e0140291. doi: 10.1371/journal.pone.0140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaney JE, Hanson CT, Hanley KA, Murphy BR, Whitehead SS. 2004. Vaccine candidates derived from a novel infectious cDNA clone of an American genotype dengue virus type 2. BMC Infect Dis 4:39. doi: 10.1186/1471-2334-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gubler DJ, Reed D, Rosen L, Hitchcock JR. 1978. Epidemiologic, clinical, and virologic observations on dengue in the Kingdom of Tonga. Am J Trop Med Hyg 27:581–589. [DOI] [PubMed] [Google Scholar]
- 72.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. 2000. Modulation of Dengue virus infection in human cells by alpha, beta, and gamma interferons. J Virol 74:4957–4966. doi: 10.1128/JVI.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. 2013. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.