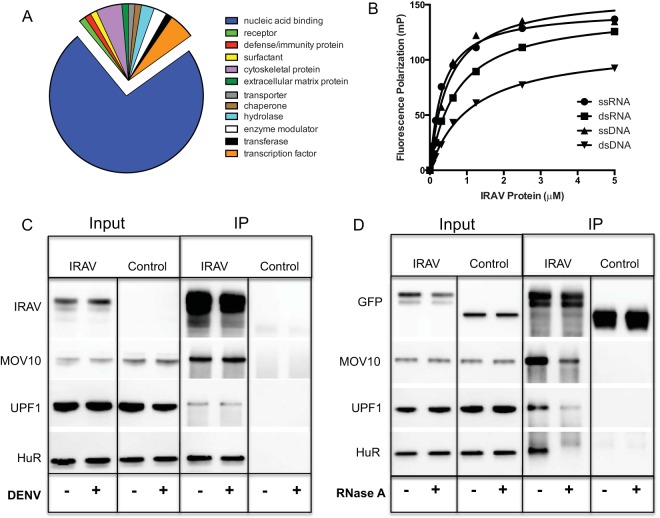
FIG 4.
IRAV interacts with RNA binding proteins. (A) Gene ontology analysis of IRAV interaction partners as identified by co-IP of GFP-IRAV from DENV-infected HEK293 cells, followed by MS analysis. Gene ontology analysis was performed using the PANTHER classification system (http://www.pantherdb.org), based on the protein class. (B) FP assays were performed using various concentrations of recombinant IRAV and FAM-labeled 20-mer single-stranded or double-stranded DNA or RNA oligomers. Samples were run in triplicate, and FP was measured on a Hidex sense microplate reader (Turku, Finland). (C) Co-IP experiments performed on HEK293 cells left untreated (−) or infected with DENV (+). The cells were transfected with expression vectors for GFP-IRAV (IRAV) or GFP-CAT (Control), followed by IP with an antibody for GFP. Western blotting was performed for IRAV putative interaction partners MOV10, UPF1, and HuR. (D) Co-IP experiments were performed on HEK293 cells after overexpression of GFP-IRAV (IRAV) or GFP-CAT (Control). Cell lysates were either left untreated (−) or treated with RNase A (+), followed by IP with an antibody for GFP. Western blots were then performed for IRAV putative interaction partners MOV10, UPF1, and HuR.