Abstract
We report the preparation and performance of TEM grids bearing stabilized non-fouling lipid monolayer coatings. These films contain NTA capture ligands of controllable areal density at the distal end of a flexible poly(ethylene glycol)2000 (PEG2000) spacer to avoid preferred orientation of surface-bound histidine-tagged (His-tag) protein targets. Langmuir-Schaefer deposition at 30 mN/m of mixed monolayers containing two novel synthetic lipids – 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[(5-amido-1-carboxypentyl)iminodiacetic acid]polyethylene glycolamide 2000) (NTA-PEG2000-DSPE) and 1,2-(tricosa-10′,12′-diynoyl)-sn-glycero-3-phosphoethanolamine-N-(methoxypolyethylene glycolamide 350) (mPEG350-DTPE) – in 1:99 and 5:95 molar ratios prior to treatment with a 5 min, 254 nm light exposure was used for grid fabrication. These conditions were designed to limit non-specific protein adsorption onto the stabilized lipid coating by favoring the formation of a mPEG350 brush layer below a flexible, mushroom conformation of NTA-PEG2000 at low surface density to enable specific immobilization and random orientation of the protein target on the EM grid. These grids were then used to capture His6-T7 bacteriophage and RplL from cell lysates, as well as purified His8-green fluorescent protein (GFP) and nanodisc solubilized maltose transporter, His6-MalFGK2. Our findings indicate that TEM grid supported, polymerized NTA lipid monolayers are capable of capturing His-tag protein targets in a manner that controls their areal densities, while efficiently blocking non-specific adsorption and limiting film degradation, even upon prolonged detergent exposure.
Keywords: affinity grid, lipid monolayer, affinity purification, cryo-electron microscopy (cryo-EM), monolayer purification, 2D polymerization
Graphical abstract
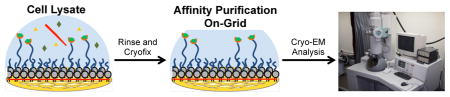
Introduction
Cryogenic electron microscopy (cryoEM) single particle reconstruction is a rapidly advancing method of protein structure elucidation that is now capable of producing structures with resolutions approaching 2.2 Å, provided that thousands of single particle images can be collected.1 At present, this is achieved by collecting and class averaging images from many different carbon-coated grid preparations that randomly adsorb and present the protein target in multiple orientations for cryoEM imaging. Since many protein target candidates exist in low copy number, may be highly labile, and/or are derived from infectious agents that are in low supply, there is significant motivation for the development of new sample preparation methods to improve the throughput and predictability of this technique.
Lipid monolayers have been used to concentrate proteins at interfaces for structure determination, as first described by Uzgiris and Kornberg.2 Subsequent affinity monolayer developments explored a variety of affinity lipid-ligand interactions such as biotin-streptavidin, ATP lipid-ATP binding protein, Ni2+:nitrilotriacetic acid (NTA)-hexahistidine-tagged (His6-tag) protein, or Cu2+:iminodiacetic acid-His6-tag protein.3 Many of these efforts were focused on the development two-dimensional crystallization methods using nitrilotriacetic acid (NTA)-modified lipid conjugates,4–9 with some cases showing the potential of NTA lipids for structure elucidation with resolutions in the 10 – 25Å regime.5 NTA lipid monolayers were extended to the realm of cryoEM single particle reconstruction by Walz and coworkers, with the structures for 50S ribosomal subunits,10 transferrin-transferrin receptor complex,11, 12 and human RNA polymerase II13 reported using a commercially available NTA lipid conjugate, 1,2-dioleoyl-sn-glycero-3-[((N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (DGS-NTA), in mixed monolayers with DLPC. They also extended this technique to the retrieval of protein targets from cell lysates;11, 13 however, the potential of the approach is currently limited by the low stability,10 non-specific fouling,11 and preferred orientation14 limitations of DGS-NTA:DLPC monolayers.
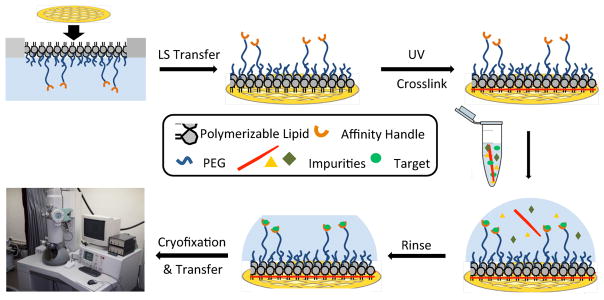
We report our attempts to address these challenges by depositing non-fouling PEG-lipid15–17 coatings containing common affinity ligands like NTA3, 8, 9, 18, 19 onto TEM grids.20 These coatings are prepared by compression of lipid films whose NTA surface density and PEG mushroom-brush conformational state can be controlled by initial film composition and applied surface pressures prior to deposition onto the EM grid. Brush conformation methoxypolyethylene glycol (mPEG) coatings are highly non-fouling,21–23 thereby enabling the application of cell lysates directly onto the grid and removal of the non-specifically bound material in subsequent buffer washing steps. In addition to the reduced sample quantity demands required for cryoEM analysis, the shortened sample processing times also limit the opportunities for proteolytic degradation and/or protein unfolding of the His6-target that may occur during standard sample processing protocols. Langmuir-Schaefer deposition of a mixed monolayer containing two synthetic polymerizable lipids – 1,2-(tricosa-10′,12′-diynoyl)-sn-glycero-3-phosphoethanolamine-N (methoxypolyethylene glycolamide 350) (mPEG350-DTPE) and 1,2-(tricosa-10′,12′-diynoyl)-sn-glycero-3-phosphoethanolamine-N-[(5-amido-1-carboxypentyl)iminodiacetic acid]polyethylene glycolamide 2000) (NTA-PEG2000-DTPE) – prior to treatment with a 5 min, 254 nm light exposure was used to prepare stabilized affinity monolayers supported on carbon coated EM grids. These grids were then used to capture His6-T7 bacteriophage and His6-RplL from cell lysates, as well as purified histidine-tagged green fluorescent protein (His8-GFP) and nanodisc embedded His6-MalFGK2 (Figure 1). Our findings indicate that polymerized NTA lipid monolayers are capable of capturing His6-protein targets in a manner that controls their areal densities, while efficiently blocking non-specific adsorption and limiting film degradation upon prolonged detergent exposure.
Figure 1.
Conceptual diagram showing stabilized monolayer affinity grid fabrication and utilization. (Top left) A fluid-phase mixed lipid monolayer comprised of Ni2+:NTA-PEG2000-DSPE and mPEG350-DTPE (1:99 or 5:95 mol:mol) was compressed until the mPEG350-DTPE component entered the brush regime and the diyne lipid chains were fully condensed; (Top center) the condensed lipid monolayer film was deposited onto carbon-coated TEM grids via Langmuir-Schaefer (LS) transfer; (Top right) photopolymerization of the LS film, followed by Ni2+ activation, produces a grid with a stable affinity capture coating; (Bottom right) solutions containing the His-tag protein of interest are deposited onto the coated grids, either as a clarified cell lysate or as a purified protein sample; (Bottom center) blotting and rinsing of the sample removes non-target material prior to (Bottom left) cryofixation (or negative staining) and sample imaging.
Experimental Methods
Synthesis of mPEG350-DTPE, NTA-PEG2000-DSPE, and NTA-PEG2000-DTPE
The detailed synthesis procedures for mPEG350-DTPE, NTA-PEG2000-DSPE, and NTA-PEG2000-DTPE (Figure 2) are reported in Supplementary Information.
Figure 2.
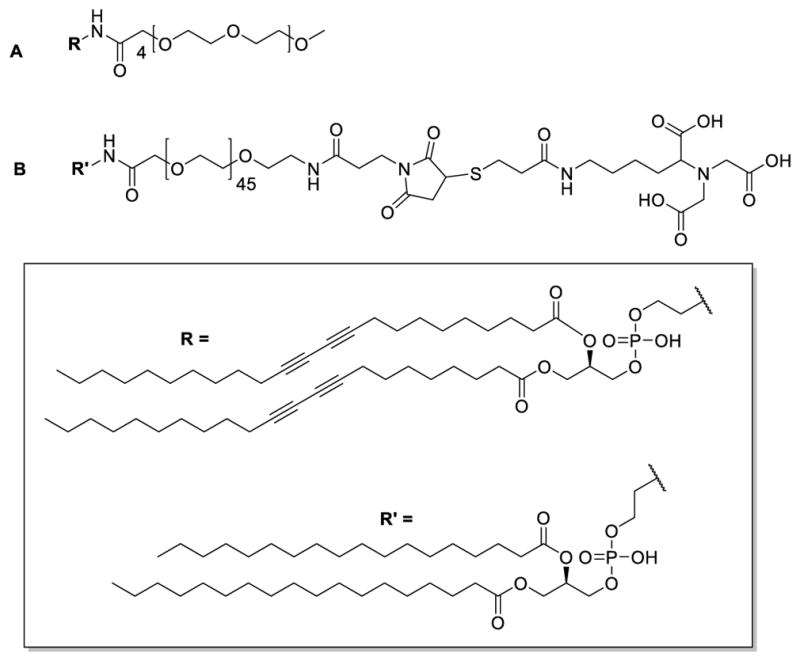
Structures of (A) mPEG350-DTPE and (B) NTA-PEG2000-DSPE.
Cell Lysate Preparations
The ASKA library, a complete E. coli K-12 ORF archive in which over 4000 bacterial proteins have been cloned into pCA24N expression vectors, was used. Each protein coding ORF has an N-terminal His6 and a C-terminal green fluorescence protein (GFP); each protein coding ORF is also available without the GFP fusion (which are the strains used in these studies). Cells containing the RplL gene overexpression vector were grown to OD = 0.6 in 100 mL of LB broth using a 37 °C shaker/incubator and induced with a final concentration of 1.0 mM IPTG before allowing to grow for an additional 4 h. After centrifugation, the supernatant was discarded before resuspending the cell pellet in lysis buffer (20 mM Tris, 10 mM MgCl2, 100 mM KCl, pH=7.4, 100 μg aprotinin, 174 μg PMSF, 500 μg lysozyme) and incubation for 20 min. The cell membranes were disrupted by sonication (35 pulses, 1 sec/pulse at 75 W) before centrifugation of the suspension at 11,000 g for 10 min. The supernatant containing His6-RplL was diluted 10-fold before application to the TEM grid for analysis.
Purified C-terminal His8-tag gp10 (capsid protein) of T7 bacteriophage was typically produced at a concentrations of ~1 × 1012 particles/mL. BL21 bacteria cells in 1 mL of LB media were grown to OD = 0.8 before addition of T7 bacteriophage (1.0 μL, 1.0×1012 particles/mL) to the culture and shaking at 37 °C for 1 h. After centrifugation of the cells, the supernatant was used directly for TEM grid analysis.
His8-GFP Purification
A glycerol stock of E. coli cells, transformed with the His8-GFPuv pT7-7 plasmid was shaken at 37 °C overnight before addition to 250 mL of LB media with shaking at 37 °C until an OD600 of 0.7 was reached. Then, a 1.0 M stock solution of isopropyl-β-D-thiogalactopyranoside (IPTG) was added, bringing the final concentration to 1.0 mM, before shaking continually at 37 °C for an additional 5 h. The cells were then centrifuged at 11,000 g for 10 min, the supernatant discarded, and the pellet re-suspended in 10 mL of lysis buffer (50 mM NaH2PO4, 100 mM NaCl, pH = 8.0, 100 μg aprotinin, 174 μg phenylmethanesulfonyl fluoride (PMSF), and 500 μg of lysozyme). The suspension was allowed to stand for 20 min before the cell membranes were disrupted using 3 rounds of sonication (35 pulses of probe sonication, 1 sec/pulse at 75 W with 1 sec rest/pulse) and the debris pelleted by centrifugation at 11,000 g for 30 min. The supernatant was incubated with 100 μL of Ni-NTA agarose beads and gently rotated for 4 h. The beads were pelleted using a bench top centrifuge operating at 5,000 g for 2 min. The supernatant was discarded and the pellet was washed with PBS buffer containing imidazole (50 mM NaH2PO4, 100 mM NaCl, 10 mM imidazole, pH = 8.0). The same spin and wash sequence was repeated with 15 mM, 20 mM, and 30 mM imidazole in PBS. The protein was eluted from the resin after a final spin, discarding of supernatant, and incubation with 500 mM imidazole in PBS for 4 h. The removal of imidazole was achieved by dialyzing the eluted protein against PBS (50 mM NaH2PO4, 100 mM NaCl, 10 mM imidazole, pH = 8.0) overnight using a 10,000 MWCO Slide-A-Lyzer. The protein was characterized using 12.5% acrylamide SDS-PAGE gel electrophoresis.
Purified Maltose Transporter Solubilized in Lipid Nanodisc
The methods used to purify and insert His6-MalFGK2 maltose transporter in lipid nanodiscs derived from membrane scaffolding proteins (MSP) followed those described in a previously reported protocol.24
Langmuir-Schaefer Film Deposition Onto TEM Grids
Stock solutions of two lipid mixtures (1:99 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE) were prepared in CHCl3 at 2.0 mg/mL and stored at −80 °C. Each of these were spread via 10 μL microsyringe at the air-water interface of a Kibron μTrough and compressed to a final surface pressure of 30 mN/m (i.e., ~59 Å2/molecule for 1:99 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers and ~66 Å2/molecule for 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers). The compressed lipid monolayers were deposited onto TEM grids by Langmuir-Schaefer transfer using tabbed TEM grids (Ted Pella) to enable grid approach at 180° (and the transfer forceps at 90°) relative to the air-water interface and the LS film immediately blotted with filter paper. (n.b.: Standard TEM grids and forceps produced extensive disruption of the interface during the LS transfer step, making it impossible to determine accurate grid transfer ratios under these conditions.) The supported monolayer films were then polymerized for 10 min using a handheld 8W/m2 254 nm lamp that was placed 6 – 8 cm above the grid surface before transfer to a standard TEM grid box for later use.
Fluorescence Microscopy
Sessile drops of His8-GFPuv solution (3.5 μL, 2.0 mg GFP/mL in PBS) were deposited onto 1:99 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE, 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE, 100% mPEG350-DTPE, or bare carbon coated grid surfaces for 2 min, followed by removal of excess sample using a microsyringe. Grids were then washed 3 times with 20 μL drops of Tris buffer before removal of the excess solution via microsyringe. For elution of Ni2+:NTA-bound T7 bacteriophage from the surface of the grid, 500 mM imidazole was deposited on the grid surface for 5 min before washing twice with 20.0 μL of PBS. After preparation, the TEM grid was sandwiched between a microscope slide and coverslip and sealed with nail polish before recording the fluorescence images.
Negative-staining Procedure
T7 bacteriophage and RplL cell lysate samples were prepared by incubating a 5.0 μL drop of cell lysate on the grid surface for 2.0 min before gentle removal by microsyringe. The grids were then washed 6 times with 20 μL drops of Tris buffer followed by a final wash with 20 μL of distilled water. A 5 μL drop of 2% uranyl acetate was placed on the grid for 1 min before removal of the excess solution with a wedge of filter paper. Ni2+:NTA-immobilized samples were eluted from the grids by adding 5 μL of 500 mM imidazole in Tris buffer immediately after sample deposition (i.e., instead of 6 Tris buffer washes). Nanodisc solubilized MalFGK2 and purified T7 bacteriophage samples were prepared in the same manner as the cell lysate samples, except that the incubation time of the samples on DLPC and mPEG350-DTPE modified grids was increased to 3 min. Negatively stained samples were imaged using a Tecnai TF20 transmission electron microscope operating at 200 kV.
Preparation of Frozen Hydrated Samples for Cryo-EM
TEM grids modified with stabilized 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers were treated with His-tag protein samples as described above for negative-staining. After application of the protein solution, the excess sample was removed by blotting the grids twice for 3 sec with an offset of −1 at 80% humidity using a Vitrobot (FEI Company). The grids were then cryo-fixed by plunging into liquid ethane and imaged at 200 kV on an FEI CM200 transmission electron microscope using low-dose techniques. Images were recorded using a Gatan Ultrascan CCD.
Results and Discussion
mPEG350-DTPE was prepared in 32% yield by treatment of commercially available 1,2-(tricosa-10′,12′-diynoyl)-sn-glycero-3-phosphoethanolamine (DTPE) with the N-hydroxysuccinimidyl (NHS) active ester form of mPEG350 (Supplementary Information, Scheme S1). NTA-PEG2000-DSPE was prepared in 9.5% yield by sequential modification of doubly activated NHS-PEG2000-maleimide with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) and a thiol-modified lysine-NTA derivative (Figure 2; Supplementary Information, Schemes S2–S4). NTA-PEG2000-DTPE was prepared in the same manner as NTA-PEG2000-DSPE, except that DTPE was used for NHS displacement instead of DSPE.
CHCl3 stock solutions of the lipid mixtures were prepared, spread at the air-water interface at 20 °C, a 5 min incubation period allowed for evaporation of the solvent, and the films compressed at a rate of 10 Å2/molecule until collapse at 30 mN/m – 35 mN/m for the mixed monolayers and 40 mN/m for pure mPEG350-DTPE monolayers. The pressure-area isotherms shown in Figure S1 for 1:99 and 5:95 NTA-PEG2000-DTPE:mPEG350-DTPE lipid mixtures displayed gradually increasing surface pressures upon film compression, except that the onset of surface pressure occurred at larger molecular areas as the NTA-PEG2000-DTPE composition in the film increased. We attribute this observation to the displacement of surface-adsorbed NTA-PEG2000 from the air-water interface as previously described25 for mixed mPEG2000-lipid monolayers. As this desorption process progresses, the majority component, mPEG350, undergoes a mushroom-brush regime transition upon compression to 30 mN/m while the NTA-PEG2000 fraction remains in the mushroom configuration. The extension of the PEG2000 polymer mushroom conformation from the surface of the film can be calculated by Lo = αN(α/D)2/3, where Lo = the length of PEG graft, α = length of PEG monomer (3.5 Å), N = number of ethylene glycol units in the PEG chain (i.e., 45), and D = the distance between grafting sites. For a lipid film containing 1% NTA-PEG2000-DTPE, the grafting site separation is estimated to be 47 Å and the Lo = 2.8 nm. Fully compressed 5:95 NTA-PEG2000-DTPE:mPEG350-DTPE monolayers are estimated to have a grafting site separation of 9.4 Å and a NTA-PEG2000 graft extension of 8.2 nm. As the lipid film is compressed below 113 Å2/molecule, the mPEG350 fraction enters the brush regime, while the NTA-PEG2000 polymer chain accommodates further compression by extending farther away from the air-water interface in a concentration-dependent manner. Consequently, a longer NTA-PEG was prepared for these studies since it would confer greater conformational freedom to the immobilized histidine-tagged target and present it in a greater diversity of possible orientations for cryoEM single particle analysis.
Since the lipid films typically collapsed at surface pressures above 30 mN/m, the monolayers were compressed to 30 mN/m prior to Langmuir-Schaefer (LS) transfer onto carbon-coated grids that were not subjected to glow discharge before LS deposition. This method produced transfer ratios between 0.89 – 0.97 for the LS deposition step. Photopolymerization of the lipid monolayer was initiated by placing the grids ~6 – 8 cm from a handheld UV lamp and irradiated for 10 min. Grid performance as a function of varied irradiation time suggested that a 10 min exposure was most favorable.
After monolayer film transfer onto 400 mesh grids and staining with freshly prepared 1% UO2(OAc)2, TEM images revealed the presence of large continuous areas exceeding 900 μm2 in size that were punctuated by much smaller zones of higher contrast (Figure S2). We infer from these findings that the large area regions were monolayer films with occasional, interspersed domains of bilayer and trilayer films arising from partial monolayer collapse during compression that were preserved by the photopolymerization process. Sufficiently large areas of the grid were coated by stabilized monolayer film structures to make them suitable for negative stain and cryoEM studies; however, the multi-layer sections of the film were helpful for monitoring the stability of the film in the presence of detergents as described below.
The anti-fouling properties of the mPEG350 brush layer within the stabilized monolayer coating was probed qualitatively using fluorescence microscopy analysis of His8-GFPuv adsorption onto these modified surfaces. Wide field and line scan fluorescence microscopy analysis revealed that glow discharged bare amorphous carbon surfaces possessed nearly 20-fold greater fluorescence intensities after exposure to His8-GFPuv than grids bearing mPEG350 brush regime coatings (>8000 RFU and ~400 RFU, respectively; Figure S3). Our data shows that the fluorescence distribution was uniform over both types of surfaces, however, in rare instances there were small variations in fluorescence observed. In those cases, we attribute the areas of decreased fluorescence to grid regions with an incomplete carbon coating, whereas zones of substantially increased fluorescence were ascribed to incomplete LS transfer of the brush layer mPEG350-DTPE. We infer from these findings that non-specific binding of protein on the brush regime mPEG350-DTPE coated grids is much lower than that of glow discharged bare carbon films due to the combined effects of molecular weight, polymer hydration, excluded volume, elastic restoration, and grafting density on the surface that enhance the antifouling properties of the mPEG coating.22, 26, 27 In some cases, low molecular weight PEG polymers such as mPEG350 display greater antifouling properties than their higher molecular weight counterparts since the shorter PEG chains are better able to form a more densely packed layer.28, 29 Based on these findings, we concluded that brush regime mPEG350-DTPE monolayers, deposited onto a carbon coated surface by LS transfer, would be a good candidate for blocking non-specific adsorption of macromolecules onto EM grid surfaces. To test this assumption, we conducted a comparative study between DLPC monolayers and mPEG350-DTPE stabilized monolayers, both deposited via LS transfer, using bare carbon grids as a control.
His8-GFPuv capture on grids bearing stabilized 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers was first demonstrated by fluorescence microscopy. Wide-field fluorescence images (Figure S4) of Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE modified grids reveal a non-uniform, concentration-dependent GFP fluorescence. Some spots of bright fluorescence, suggestive of GFP aggregation, were also observed. The observed pixel intensities in the fluorescence line scan analyses were higher for the grids bearing a 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE coating than the grids coated with a 1:99 monolayer. In both cases, the bound His8-GFPuv could be removed from the grid surface after a brief 5 min exposure to either 500 mM imidazole or 150 mM EDTA to remove Ni2+ from the His8:Ni2+:NTA complex to enable elution of the immobilized protein target from stabilized, Ni2+ activated 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers. These findings indicate that His-tag targets can be captured in an NTA concentration-dependent manner, with higher NTA loadings producing greater target protein concentrations on TEM coated with stabilized, Ni2+-activated NTA-PEG2000-DTPE:mPEG350-DTPE monolayers.
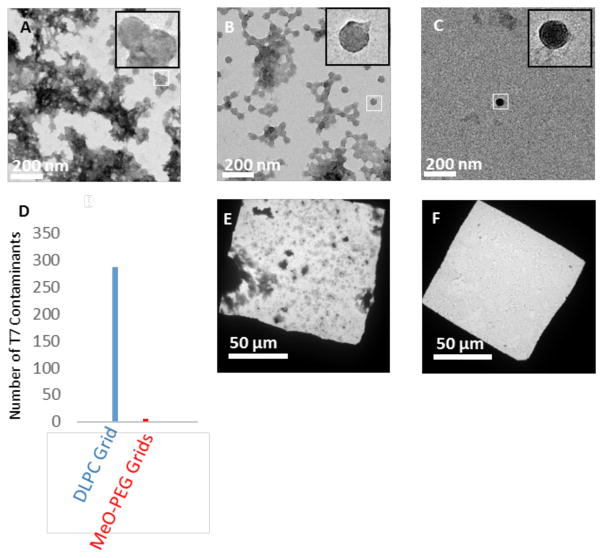
Purified His6-T7 bacteriophage (1011 – 1012 particles/mL) was used to probe the non-specific adsorption properties of DLPC and mPEG350-DTPE stabilized monolayer-coated grids at higher resolution, with carbon coated, glow-discharged grids serving as a control. The sample precipitated on grids lacking modified DLPC or MPEG350 monolayers (Figure 3A). We identified virus-like particles on these grids, as shown in Fig 3A (inset); however the poor sample quality produced on unmodified, glow-discharged grids made accurate quantification of adsorbed virus particles impossible. Therefore, we only quantified virus adsorption onto DLPC and mPEG350 modified grids, as discrete particles could be discerned in both cases. DLPC coated grids were found to decrease the amount of T7 that was adsorbed to the grid relative to bare carbon grids (Figure 3B), indicating that interactions between T7 bacteriophage and the DLPC monolayer are not as favorable. Nonetheless, a significant amount of occult debris remained on the DLPC monolayer coated grids. Grids coated with mPEG350-DTPE stabilized monolayers; however, displayed a greatly reduced accumulation of both T7 and debris (Figure 3C). The appearance of the virus particles in these samples included round-like objects as well as more familiar hexagonal projection of the mature icosahedral form of the virus. The T7 phage particles in these images represent the immature form where an absence of mature capsid shell protein gives results in the rounded appearance of these particles.30 The low abundance of T7 phage particles highlights the antifouling opportunities of these PEGylated surfaces. Quantitative comparisons of the T7 particle densities on DLPC and mPEG350 monolayers revealed an accumulation of ~110 particle/1 μm2, and 5 particle/1 μm2, respectively. We infer from these findings that mPEG350-DTPE stabilized monolayers are the most effective toward blocking non-specific adsorption of T7 bacteriophage (Figure 3D). Low magnification images of purified T7 deposited onto both DLPC and mPEG-DTPE stabilized monolayer coated grids reveal a substantially lower background in the case of mPEG350-DTPE coated grids (Figure 3E and 3F).
Figure 3.
Comparative performance of uncoated (bare carbon, glow-discharge), DLPC, and mPEG350-DTPE coated grids toward non-specific protein adsorption (i.e., lacking NTA groups to promote specific binding). The capacity of these grids to reject non-specific protein adsorption was tested by negative stain TEM analysis using His6-T7 bacteriophage in purified form or within cell lysates. (A) Purified His6-T7 bacteriophage on glow-discharged bare carbon grid; (B) purified His6-T7 bacteriophage on 100% DLPC monolayer coated grid; (C) His6-T7 bacteriophage on stabilized 100% mPEG350-DTPE monolayer coated grid; (D) number of non-specific contaminants adsorbed from purified His6-T7 bacteriophage solution onto bare carbon grids (blue), DLPC coated grids (red), and mPEG350-DTPE coated grids (green) as determined by counting 60 randomly selected fields across 3 different grids for each grid type; (E) His6-T7 bacteriophage in cell lysate applied to 100% DLPC coated grid; and (F) His6-T7 bacteriophage in cell lysate applied to stabilized 100% mPEG350-DTPE coated grid.
After evaluating the capacity of these grids to inhibit non-specific adsorption, we next turned our attention to their ability to direct affinity-guided interactions with NTA-modified grids. First, we used fluorescence microscopy to monitor His8-GFPuv capture on grids bearing stabilized 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers. Wide-field fluorescence images (Figure S4) of Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE modified grids reveal a non-uniform, concentration-dependent GFP fluorescence. Occasional spots of bright fluorescence, suggestive of GFP aggregation, were also observed. The observed average pixel intensities in the fluorescence line scan analyses were higher for the grids bearing a 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE coating than the grids coated with a 1:99 monolayer. In both cases, the bound His8-GFPuv could be removed from the grid surface after a brief 5 min exposure to either 500 mM imidazole or 150 mM EDTA to remove Ni2+ from the His8:Ni2+:NTA complex to enable elution of the immobilized protein target from stabilized, Ni2+ activated 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers. These findings indicate that histidine-tagged targets can be captured in an NTA concentration-dependent manner, with higher NTA loadings producing greater target protein concentrations on TEM grids coated with stabilized, Ni2+-activated NTA-PEG2000-DTPE:mPEG350-DTPE monolayers.
Affinity capture of His6-T7 bacteriophage from cell lysates was then tested by negative stain and cryoelectron microscopy using grids modified with mPEG350-DTPE stabilized monolayers, Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers, and Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers rinsed with 500 mM imidazole after lysate exposure (Figure 4). In the presence of mPEG350-DTPE stabilized monolayers, we observed minimal binding of His6-T7 bacteriophage and very low levels of debris from the lysate onto the grid (Figure 4A). Background adsorption of non-T7 phage debris is highlighted in the inset of this image. In some cases, T7 particles were not observed on the brush polymer surface even at low magnification, suggesting a complete blockage of non-specific adsorption. Grids bearing Ni2+-charged 1:99 NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers were capable of capturing His6-T7 bacteriophage from cell lysate while limiting the background adsorption of undesired non-target cellular components (Figure 4B). It is worth noting that T7 viral particles often give mixed results of negative-stain and positive-stained particles since UO22+ salts may positively stain packaged nucleic acid within the capsid structure.31 Exposure of these samples to 500 mM imidazole led to greatly decreased surface adsorption of His6-T7 bacteriophage, suggesting that the interaction of phage particles with the supported NTA-lipid monolayer is Ni2+ specific (Figure 4C). We attribute the occurrence of the few remaining particles to either non-specific adsorption to uncoated grid regions or particles that are avidly bound due to multivalent interactions between the phage particles and the NTA-PEG grafted surface. To compensate for particle capture efficiency limitations in samples prepared for cryogenic imaging10 due to unfavorable kinetics and/or modest ligand-receptor affinities,32 we examined 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayer coated grids to determine whether the areal density of NTA ligands had a detectable influence on His6-T7 bacteriophage capture efficiency. Using identical sample processing methods and incubation times, we qualitatively observed an increase in particle surface density with increasing Ni2+:NTA-PEG2000-DTPE composition (unpublished data). Since this comparison was not performed in a side-by-side manner between 1:99 and 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers with the same lysate, it is conceivable that these findings may be the result of different virus titers in the His6-T7 bacteriophage preparations. Nonetheless, these findings are consistent with the His8-GFPuv experiments described above (Figure S4) showing that higher surface concentrations of affinity capture lipid produces greater areal densities of immobilized target on the NTA-modified surface, with the 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayer coated grids being more suitable for cryoEM sample preparation (Figure 4D). The immobilization of both mature and immature T7 phage from lysate samples (Figure S8) highlights the potential of rapid purification using affinity grids towards studying dynamic processes, which may not be possible when visualizing purified samples generated using standard TEM sample preparations.
Figure 4.
Effect of NTA surface density on His-T7 bacteriophage captured from cell lysates using negative stain and cryoEM analysis. (A) Negative stain TEM appearance of grid coated with stabilized 100% mPEG350-DTPE monolayer after 2 min exposure to cell lysate containing His-T7 bacteriophage; (B) same as in (A), except that the grid was coated with stabilized 1:99 Ni2+:NTA-PEG2000-DSPE:mPEG350-DTPE monolayer; (C) same as in (B), except that the grid was rinsed with 500 mM imidazole, pH = 7.4 after the 2 min lysate exposure step; and (D) cryoEM appearance of grid coated with stabilized 5:95 Ni2+:NTA-PEG2000-DSPE:mPEG-350-DTPE monolayer after 2 min exposure to cell lysate containing His-T7 bacteriophage.
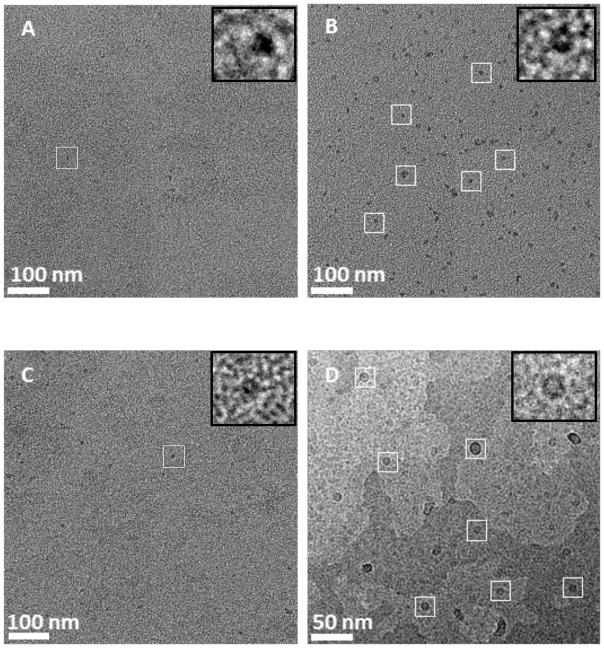
We next demonstrated the capture of the 50S E. coli ribosomal subunit,10 His6-RplL, directly from cell lysates onto grids coated with Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers. Negative stain EM images of His6-RplL lysate deposited onto grids coated with mPEG350-DTPE stabilized monolayers showed no appreciable target-specific adsorption onto the grid (Figure 5A). After LS deposition of 20:80 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers onto the grids, particles consistent with the size and various shapes of the 50S subunit were observed on the grid surface (Figure 5B). When 500 mM imidazole was added to samples containing the captured ribosomal particles on 20:80 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers, the previously adsorbed particles were eluted from the surface of the TEM grid (Figure 5C), consistent with the expectation that the interaction of His6-RplL with the surface of the grid is Ni2+ specific. Using the same sample preparation procedure, cryoEM analysis revealed the capture of His6-RplL from E. coli lysates (Figure 5D) as crown-like projections in some images (Figure 5D). These have frequently been reported and are attributed to the 50S ribosomal subunit. The earliest reported TEM images of ribosomes were small, featureless bumps on the grid surface;33 however, more powerful tools eventually led to the revelation of several features in the ribosomal units such as ‘crown-like’ and ‘duck-like’ projections for the 50S and 30S subunits, respectively.34, 35 Since the RplL protein carries the hexahistidine tag on the 50S subunit in our construct, we identified many more of these particles bound to the grid than the 30S subunit. Crown-like projections are clearly seen from the side view of the 50S particle with a central protuberance, a ridge, a stalk, and two groove-like features.
Figure 5.
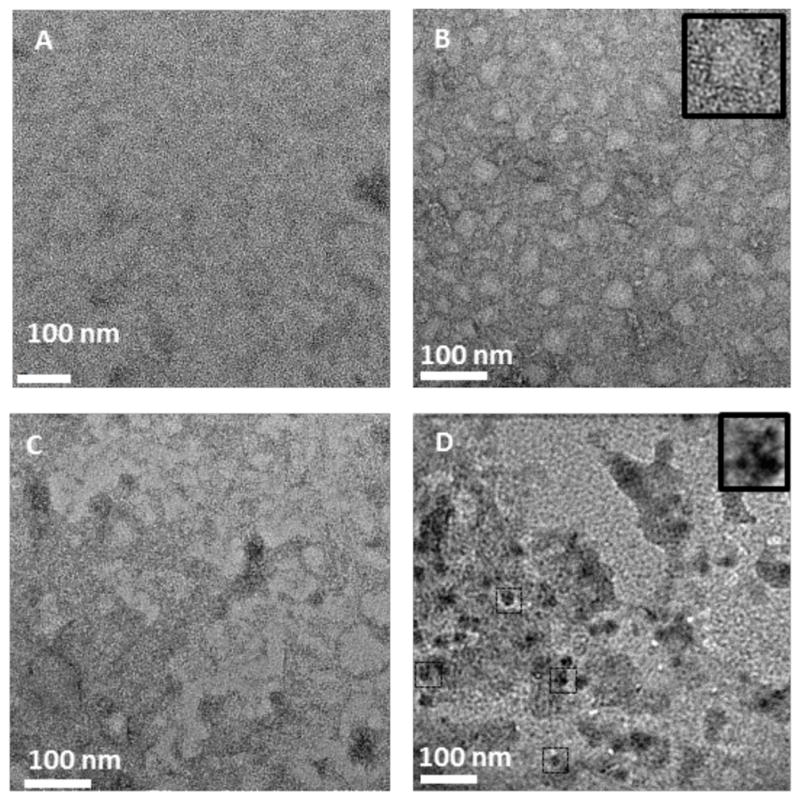
Affinity capture of His-RplL from cell lysates onto stabilized Ni2+:NTA-PEG2000-DTPE:mPEG-350-DTPE monolayer coated grids. (A) Negative stain TEM of grid coated with stabilized 100% mPEG350-DTPE after 2 min exposure to cell lysate containing His-RplL; (B) negative stain TEM of grid coated with 20:80 Ni2+:NTA-PEG2000-DSPE:mPEG350-DTPE after 2 min exposure to cell lysate containing His-RplL; (C) same as in (B), except that the grid was rinsed with 500 imidazole, pH=7.4 after the 2 min exposure to cell lysate containing His-RplL; and (D): cryoEM image of grid coated with stabilized 5:95 Ni2+:NTA-PEG2000-DSPE:mPEG-350-DTPE monolayer after 2 min exposure to cell lysate containing His-RplL.
A key design feature of these stabilized affinity coatings is their utilization of two different types of poly(ethylene glycol)-modified lipids, one high molar ratio brush layer short segment to block non-specific adsorption and facilitate stabilization via photopolymerization (i.e., mPEG350-DTPE) and a second low molar ratio component to enable multiple orientations of the captured protein target to avoid preferred orientations at the monolayer interface and provide a more representative single particle reconstruction analysis data set. We tested this hypothesis by preparing dozens of TEM grids bearing stabilized Ni2+:NTA-PEG2000-DSPE:mPEG350-DTPE coatings that had been treated with His6-RplL containing lysates. CryoEM evaluation of these grids confirmed that the NTA-PEG2000 tether is capable of both capturing and presenting multiple orientations of the bound His6-RplL target as demonstrated by the overlay of the known 50S subunit and 70S complex structures in different rotational configurations (data not shown).
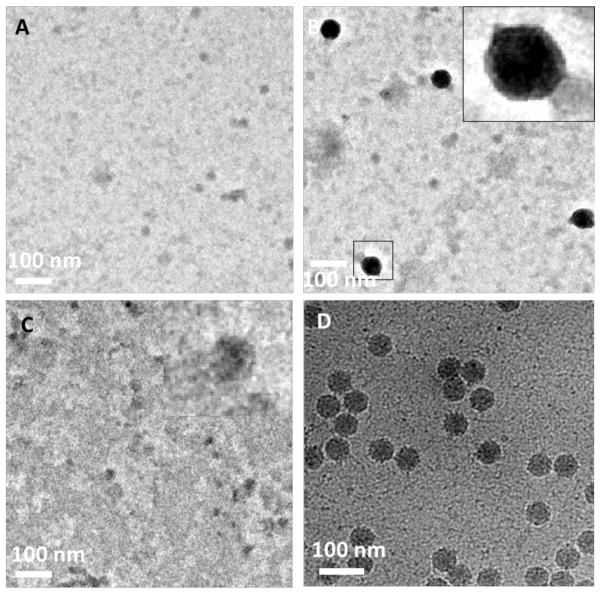
MSP have become an increasingly popular tool for stabilizing membrane protein dispersions since it provides a platform in which the local environment of solubilized membrane proteins resemble the natural environment of the lipid bilayer in cells.36 MSP are used to corral lipids and membrane proteins within disk-like structures containing a lipid bilayer interior that can host reconstituted membrane proteins for structure analysis by electron microscopy,37, 38 protein modification studies,39 and protein activation analysis.40 We used the purified hexahistidine-tagged maltose transporter, His6-MalFGK2, solubilized in MSP lipid nanodisc, for evaluating their affinity capture properties on NTA-modified stabilized affinity grids. Although His-tag MSP can be used for affinity purification,41 we used a His6-MalFGK2 construct for nanodisc reconstitution to ensure that only nanodisc containing His6-MalFGK2 are surface immobilized. We found that mPEG350-DTPE stabilized monolayer coated grids treated with His6-MalFGK2 nanodisc appeared to have fewer particles bound to the grid surface (Figure 6A) than the grids coated with 1:99 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers (Figure 6B). Treatment of the His6-MalFGK2-immobilized grids with 500 mM imidazole produced a decrease in MSP nanodisc binding to the surface (Figure 6C), further suggesting that the interaction between nanodisc solubilized His6-MalFGK2 and the NTA-modified surface is Ni2+ specific. High magnification images of nanodisc solubilized His6-MalFGK2 immobilized onto 5:95 Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE stabilized monolayers (Figure 6D) reveal a set of disc-like features in the size range of other lipid nanodisc dispersions (7 – 13 nm in diameter).42 Although the His6-MalFGK2 within the MSP nanodiscs is not directly observed due to its small size relative to the nanodiscs, the fact that these features elute with 500 mM imidazole strongly corroborates the assumption that they contain His6-MalFGK2. In addition, our images show predominantly top/bottom views of the particles, i.e., a disk-like appearance in TEM micrographs (Figure 6D), suggesting that a longer PEG spacer is needed to collect a greater number of side views for this solubilized target.
Figure 6.
Affinity capture of MSP nanodisc containing purified His-MalFGK2. (A) Negative stain TEM of stabilized 100% mPEG350-DTPE monolayer-coated grid after treatment with His-MalFGK2 in nanodiscs; (B) negative stain TEM of His-MalFKG2 in nanodisc captured on stabilized 1:99 Ni2+:NTA-PEG2000-DSPE:mPEG350-DTPE monolayer-coated grid; (C) same as in (B), except that the grid was rinsed with 500 mM imidazole, pH = 7.4 after the His-MalFGK2 in nanodisc exposure step; and (D) cryoEM of His-MalFGK2 in nanodisc captured on stabilized 5:95 Ni2+:NTA-PEG2000-DSPE:mPEG-350-DTPE monolayer-coated grid.
In order to test the durability of the stabilized monolayers toward detergent exposure, the modified grids were treated with either 0.03% Triton X-100 (CAC = 0.015%), 0.014% Tween 20 (CAC = 0.0072%), and 0.5% CHAPS (CMC=0.49%) solutions for varying periods of time before TEM observation. Lipid monolayers adsorbed to solid substrates display better detergent resistance than lipid bilayers that are prone to disruption via lipid flip-flop.43, 44 Initial encounter of detergents with the outer bilayer leaflet results in fluidization and lipid flip-flop from the inner leaflet to the outer leaflet, eventually leading to complete solubilization of the bilayer structure.44 Inhibition of flip-flop due to adsorption of a highly compressed monolayer onto the solid carbon substrate inhibits flip-flop processes, leading to improved detergent resistance.
Despite some evidence of enhanced detergent resistant of supported DLPC monolayers, prior attempts to produce TEM grids coated with mixed Ni2+:NTA-DSG:DPLC monolayers showed that their exposure to harsher detergents such as Triton X-100, Tween-20, and CHAPS led to monolayer solubilization.10 We tested the detergent resistance of our stabilized Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayer coatings under conditions where DLPC monolayers failed by using 0.03% Triton X-100 (CMC = 0.015%), 0.014% Tween 20 (CMC = 0.0072%), and 0.5% CHAPS (CMC=0.49%) at 5, 15, and 30 min exposures to the detergent solutions. Negative stain TEM observation of the coatings after detergent exposure indicated that stabilized Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayer coatings remain intact after 30 minutes of incubation with Triton X-100, Tween-20, and CHAPS (Figure S6). In the case of Triton X-100, some holes in the monolayer were occasionally observed, but none were apparent in grids treated with Tween 20 or CHAPS. The capacity of these coatings to immobilize purified His6-T7 bacteriophage after detergent exposure for 20 min indicated that a modest reduction in phage binding to the grid occurred, however, the immobilized His6-T7 bacteriophage particles were bound specifically as indicated by their ability to be removed by elution with 500 mM imidazole (unpublished data).
Fluorescence microscopy analysis of detergent-treated Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayers revealed that the stabilized Ni2+ lipid monolayer coatings retained their ability to capture His8-GFPuv. Pixel intensity data was collected over the entire fluorescence image (Figure 7) to best represent the overall specific capture capacity of the coating, regardless of whether it had been detergent compromised or not. In the case of Triton X-100 exposed samples, the fluorescence was occasionally enhanced near the grid edges, possibly as a result of solubilized or damaged monolayer near the center of the grid holes that were unable to capture His8-GFPuv in those regions. We did not see this phenomenon with either CHAPS- or Tween 20-treated surfaces. These grids exhibited a uniform distribution of His8-GFPuv fluorescence over the grid holes until the surfaces were treated with 500 mM imidazole to displace the immobilized protein. Despite the limitations of these stabilized Ni2+:NTA-PEG2000-DTPE:mPEG350-DTPE monolayer coatings toward prolonged Triton X-100 exposure, our data indicates that histidine-tagged proteins can be reversibly immobilized even after detergent exposure. We infer from these findings that both the integrity of the stabilized lipid monolayer and the specific affinity capture of the grids remains intact after exposure to detergents that has proven detrimental to previously reported mixed Ni2+:NTA-DGS:DLPC monolayers.10
Figure 7.
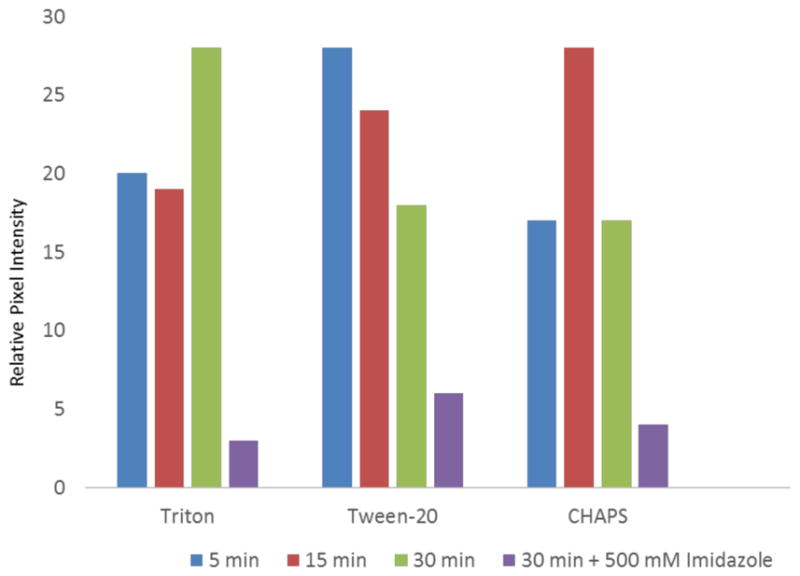
Effect of detergent exposure on 1:99 Ni2+:NTA-PEG2000-DSPE:mPEG350-DTPE stabilized monolayer affinity grids as determined by fluorescence microscopy using His-GFP as a probe for NTA monolayer retention.
Conclusions
Langmuir-Schaefer transfer of compressed, mixed lipid monolayers containing Ni2+:NTA-PEG2000:mPEG350 headgroups in 1:99 and 5:95 molar ratios are capable of capturing His-tag protein targets from cell lysates with controllable areal densities and low degrees of non-specific protein adsorption. Photopolymerization of the mPEG350-DTPE component in these monolayers by irradiating for 5 min at 254 nm produced EM grids with stabilized lipid coatings that were used to then used to capture His6-T7 bacteriophage and His6-RplL from cell lysates, purified His8-GFPuv, and nanodisc embedded His6-MalFGK2. Our data indicate that the crosslinked lipid coating maintains the non-fouling properties of the monolayer and provides multiple orientations of the captured target on the surface, while also affording greater mechanical and detergent resistance than previously reported for EM grid-supported DLPC monolayers.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Amy L. Davidson. The financial support of NIH Grant #R41GM098017 is gratefully acknowledged.
References
- 1.Bartesaghi A. 2.2 A resolution cryo-EM structure of [beta]-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uzgiris EE, Kornberg RD. Two-dimensional Crystallization Technique for Imaging Macromolecules, With Application to Antigen-Antibody Complement Complexes. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 3.Thompson DH, Zhou M, Grey J, Kim H-k. Design, Synthesis and Performance of NTA-Modified Lipids as Templates for Histidine-Tagged Protein Crystallization. Chemistry Lett. 2007;36:956–975. [Google Scholar]
- 4.Schmitt L, Dietrich C, Tampe R. Synthesis and Characterization of Chelator-lipids for Reversible Immobilization of Engineered Proteins at Self-assembled Lipid Interfaces. J Am Chem Soc. 1994;116:8485–8491. [Google Scholar]
- 5.Kubalek EW, Le Grice SFJ, Brown PO. Two-dimensional Crystallization of Histidine-tagged, HIV-1 Reverse Transcriptase Promoted by a Novel Nickel-chelating Lipid. J Struct Biol. 1994;113:117–123. doi: 10.1006/jsbi.1994.1039. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich C, Schmitt L, Tampe R. Molecular Organization of Histidine-tagged Biomolecules at Self-assembled Lipid Interfaces Using a Novel Class of Chelator Lipids. Proc Nat’l Acad Sci USA. 1995;92:9014–9018. doi: 10.1073/pnas.92.20.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich C, Boscheinen O, Scharf KD, Schmitt L, Tampe R. Functional Immobilization of a DNA-binding Protein at a Membrane Interface via Histidine Tag and Synthetic Chelator Lipids. Biochemistry. 1996;35:1100–1105. doi: 10.1021/bi952305+. [DOI] [PubMed] [Google Scholar]
- 8.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M, Fuller S, Karanjia S, Love Z, Jones R, Zhao X, Rui Y, Thompson DH. Structural Analysis of Membrane-Bound Retrovirus Capsid Proteins. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barklis E, McDermott J, Wilkens S, Fuller S, Thompson DH. Organization of HIV-1 Capsid Proteins on a Lipid Monolayer. J Biol Chem. 1998;273:7177–7180. doi: 10.1074/jbc.273.13.7177. [DOI] [PubMed] [Google Scholar]
- 10.Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: A Pre-fabricated EM grid for Monolayer Purification. J Mol Biol. 2008;382:423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly DF, Dukovski D, Walz T. Monolayer Purification: A Rapid Method for Isolating Protein Complexes for Single-particle Electron Microscopy. Proc Nat’l Acad Sci USA. 2008;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dukovski D, Li Z, Kelly DF, Mack E, Walz T. Structural and Functional Studies on the Stalk of the Transferrin Receptor. Biochem Biophys Res Comm. 2009;381:712–716. doi: 10.1016/j.bbrc.2009.02.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly DF, Dukovski D, Walz T. Strategy for the Use of Affinity Grids to Prepare Non-His-Tagged Macromolecular Complexes for Single-Particle Electron Microscopy. J Mol Biol. 2010;400:675–681. doi: 10.1016/j.jmb.2010.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma G, Pallesen J, Das S, Grassucci R, Langlois R, Hampton CM, Kelly DF, des Georges A, Frank J. Affinity Grid-based Cryo-EM of PKC Binding to RACK1 on the Ribosome. J Struct Biol. 2013;181:190–194. doi: 10.1016/j.jsb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boomer JA, Inerowicz HD, Zhang Z-Y, Bergstrand N, Edwards K, Kim J-M, Thompson DH. Acid-Triggered Release from Sterically-Stabilized Fusogenic Vesicles: A Novel DePEGylation Strategy. Langmuir. 2003;19:6408–6415. [Google Scholar]
- 16.Longo G, Thompson DH, Szleifer I. Ligand-Receptor Interactions Between Surfaces: The Role of Binary Polymer Spacers. Langmuir. 2008;24:10324–10333. doi: 10.1021/la8009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin J, Shum P, Grey J, Malhotra GS, Fujiwara S, González-Bonet AM, Moase E, Allen TM, Thompson DH. Acid-labile PEG-Vinyl Ether-Lipids with Tunable pH Sensitivity: Synthesis and Structural Effects on Hydrolysis Rates, Calcein Release Performance and Biodistribution of Their DOPE Dispersions. Mol Pharm. 2012;9:3266–3276. doi: 10.1021/mp300326z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M, Haldar S, Franses J, Kim J-M, Thompson DH. Synthesis and Self-Assembly Properties of Acylated Cyclodextrins and Nitrilotriacetic Acid (NTA)-Modified Inclusion Ligands for Interfacial Protein Crystallization. Supramolecular Chem. 2005;17:101–111. [Google Scholar]
- 19.Kang E, Park J-w, McClellan S, Kim J-M, Holland DP, Lee GU, Franses E, Park K, Thompson DH. Specific Adsorption of Histidine-Tagged Proteins on Silica Surfaces Modified with Ni2+:NTA-Derivatized Poly(ethylene glycol) Langmuir. 2007;23:6281–6288. doi: 10.1021/la063719e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Vago F, Zhang D, Snyder JE, Yan R, Zhang C, Benjamin C, Jiang X, Kuhn RJ, Serwer P, Thompson DH, Jiang W. Single-step Antibody-based Affinity Cryo-Electron Microscopy for Imaging and Structural Analysis of Macromolecular Assemblies. J Struct Biol. 2014;187:1–9. doi: 10.1016/j.jsb.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JH, Kopecek J, Andrade JD. Protein-resistant Surfaces Prepared by PEO-containing Block Copolymer Surfactants. J Biomed Mater Res. 1989;23:351–368. doi: 10.1002/jbm.820230306. [DOI] [PubMed] [Google Scholar]
- 22.Jeon SI, Andrade JD. Protein-Surface Interactions in the Presence of Polyethylene Oxide. J Coll Interfac Sci. 1991;142:159–166. [Google Scholar]
- 23.Malmsten M, Emoto K, Van Alstine JM. Effect of Chain Density on Inhibition of Protein Adsorption by Poly(ethylene glycol) Based Coatings. J Coll Interfac Sci. 1998;202:507–517. [Google Scholar]
- 24.Alvarez FJD, Orelle C, Davidson AL. Functional Reconstruction of an ABC Transporter in Nanodisc for use in Electron Paramagnetic Resonance Spectroscopy. J Am Chem Soc. 2010;132:9513–9515. doi: 10.1021/ja104047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boomer JA, Qualls MM, Inerowicz HD, Haynes RH, Patri VS, Kim J-M, Thompson DH. Cytoplasmic Delivery of Liposomal Contents Mediated by an Acid-labile Cholesterol-Vinyl Ether-PEG Conjugate. Bioconjugate Chem. 2009;20:47–59. doi: 10.1021/bc800239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen PL, Cohen JA, Podgornik R, Parsegian VA. Osmotic Properties of Poly(Ethylene Glycols): Quantitative Features. Biophys J. 2003;84:350–355. doi: 10.1016/S0006-3495(03)74855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ham ASW, Klibanov AL, Lawrence MB. Action at a Distance: Lengthening Adhesion Bonds with Poly(ethylene glycol) Spacers Enhances Mechanically Stressed Affinity for Improved Vascular Targeting of Microparticles. Langmuir. 2009;25:10038–10044. doi: 10.1021/la900966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen TDH, Perrin F-X, Dinh LN. New Hybrid Materials Based on Poly(etheleneoxide)-Grafted Polysilazane by Hydrosilylation and Their Anti-fouling Activities. Beilstein J. 2013;4:671–677. doi: 10.3762/bjnano.4.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee I, Pangule RC, Kane RS. Antifouling Coatings: Recent Developments in the Design of Surfaces That Prevent Fouling by Proteins, Bacteria, and Marine Organisms. Adv Mater. 2011;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 30.Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta M, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. Embo J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackermann HW, Jolicoeur P, Berthiaume L. Advantages and inconveniences of uranyl acetate in comparative virology: study of 4 tailed bacteriophages. Canadian journal of microbiology. 1974;20:1093–1099. [PubMed] [Google Scholar]
- 32.Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, Uchtenhagen H, Urlaub H, Herzog F, Peters JM, Poerschke D, Luhrmann R, Stark H. Grafix: Sample Preparation for Single-particle Electron Microscopy. Nature Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 33.Wittmann HG. Architecture of Prokaryotic Ribosomes. Annu Rev Biochem. 1983;52:35–65. doi: 10.1146/annurev.bi.52.070183.000343. [DOI] [PubMed] [Google Scholar]
- 34.Frank J, Zhu, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata RK, Agrawal RK. A Model of Protein Synthesis Based on Cryo-electron Microscopy of the E. coli Ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 35.Stark H, Mueller F, Orlova EV, Schatz M, Dube P, Erdemir T, Zemlin F, Brimacombe R, van Heel M. The 70S Escherichia coli Ribosome at 23Å Resolution: Fitting the Ribosomal RNA. Structure. 1995;3:815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- 36.Baas BJ, Denisov IG, Sligar SG. Homotropic Cooperativity of Monomeric Cytochrome P450 3A4 in a Nanoscale Native Bilayer Environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Frauenfeld J, Gumbart J, van der Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckman R. Cryo-EM Structure of the Ribosome-SecYE Complex in the Membrane Environment. Nature Struct Mol Biol. 2011;5:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandit A, Shirzad-Wasei N, Wlodarczyk LM, van Roon H, Boekema EJ, Dekker JP, de Grip WJ. Assembly of the Major Light-harvesting Complex II in Lipid Nanodiscs. Biophys J. 2011;101:2507–2515. doi: 10.1016/j.bpj.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional Reconstitution of beta2-Adrenergic Receptors Utilizing Self-assembling Nanodisc Technology. BioTechniques. 2006;40:601–612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Raines LL, Hooy RM, Roberson H, Leahy DJ, Cole PA. Tyrosine Phosphorylation of Mig6 Reduces its Inhibition of the Epidermal Growth Factor Receptor. ACS Chem Biol. 2013;8:2372–2376. doi: 10.1021/cb4005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen HH, Lithgow T, Martin LL. Reconstitution of Membrane Proteins into Model Membranes: Seeking Better Ways to Retain Protein Activities. Int J Mol Sci. 2013;14:1589–1607. doi: 10.3390/ijms14011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ly S, Bourguet F, Fischer NO, Lau EY, Coleman MA, Laurence TA. Quantifying Interactions of a Membrane Protein Embedded in a Lipid Nanodisc Using Fluorescence Correlation Spectroscopy. Biophys J. 2014;106:L5–L8. doi: 10.1016/j.bpj.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ngassam VN, Howland MC, Sapuri-Butti A, Rosidic N, Parikh AN. A Comparison of Detergent Action on Supported Lipid Monolayers and Bilayers. Soft Matter. 2012;8:3734–3738. [Google Scholar]
- 44.Kragh-Hansen U, le Maire M, Moller JV. The Mechanism of Detergent Solubilization of Liposomes and Protein-Containing Membranes. Biophys J. 1998;75:2932–2946. doi: 10.1016/S0006-3495(98)77735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.