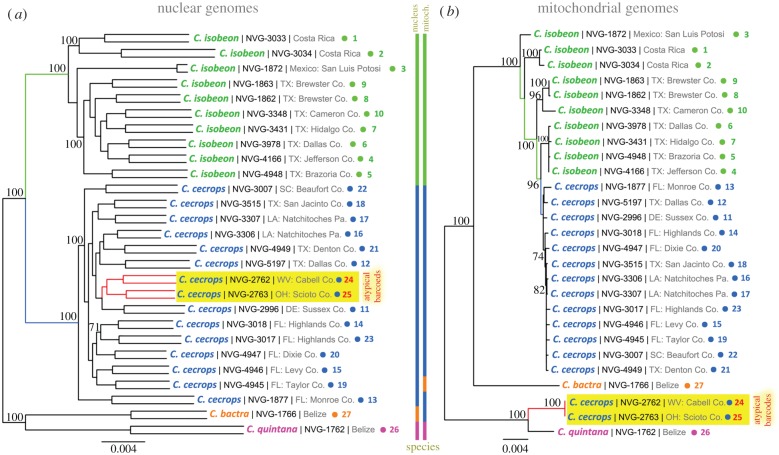
Figure 3.
Phylogenetic trees from nuclear and mitochondrial genomes of Calycopis. The trees are constructed on the concatenated alignments of the protein-coding genes in the (a) nuclear and (b) mitochondrial genomes. Both trees are unrooted. Each specimen is represented by two branches in the nuclear tree: father and mother copies of the genome (the copies are not phased and SNPs are assigned randomly to them; due to very poor coverage of specimen NVG-1872, many SNPs are impossible to call confidently and the terminal branches are very short). Name, voucher number and general locality are shown for each specimen. A number (1 to 27) points to locality of the specimen on the map shown in figure 1. Additional information about the specimens is in electronic supplementary material, table S1. Bootstrap support values above 70% are shown by the nodes, tree branches leading to C. cecrops specimens with atypical barcodes are shown in red and their names are highlighted in yellow. Coloured bars show how species are grouped in the trees. (Online version in colour.)