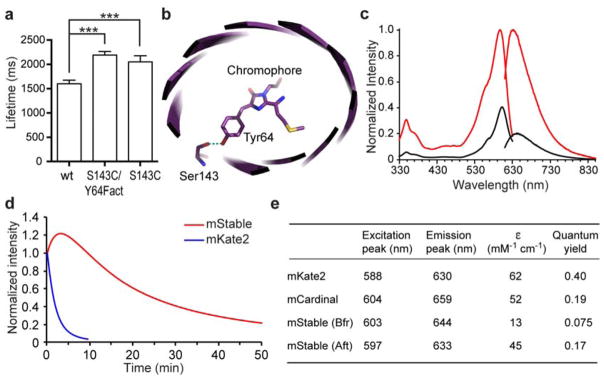
Figure 1. mStable, a far-red fluorescent protein showing unusual photostability.
(a) Single molecule measurements show that the lifetimes (mean ± s.e.) of an mKate2 mutant S143C/Y64Fact (2190 ± 73 ms, n = 364) and mutant S143C (2052 ± 125 ms, n = 37) were longer than that of the wild type mKate2 (1600 ± 74 ms, n = 193). *** P < 0.0001, unpaired t test. Fact, unnatural amino acid p-acetyl-L-phenylalanine. (b) Crystal structure of mKate (PDB ID 3BXB)20 showing Ser143 hydrogen bonding with the hydroxybenzylidene phenolate group of the chromophore. mKate2 is derived from mKate with mutations of V45A, S158A and K231R 26. (c) Fluorescence excitation and emission spectra of mStable before (black line) and after (red line) photoactivation. Emission spectra were taken at the excitation wavelength 600 nm, and emission was monitored at 640 nm for excitation spectra. (d) Photobleaching kinetics of purified mStable and mKate2 in aqueous microdroplets under arc-lamp illumination with 580/20 nm excitation filter. mKate2 t1/2 = 1.4 min; mStable t1/2 = 16.7 min. Note in this work, the t1/2s for mStable were all measured from the peak of photoactivation to 50% of the peak intensity. Each curve is the mean of three independent experiments. (e) Characteristics of mStable in comparison with mKate2 and mCardinal. Bfr and Aft indicate before and after photoactivation, respectively. ε, extinction coefficient.