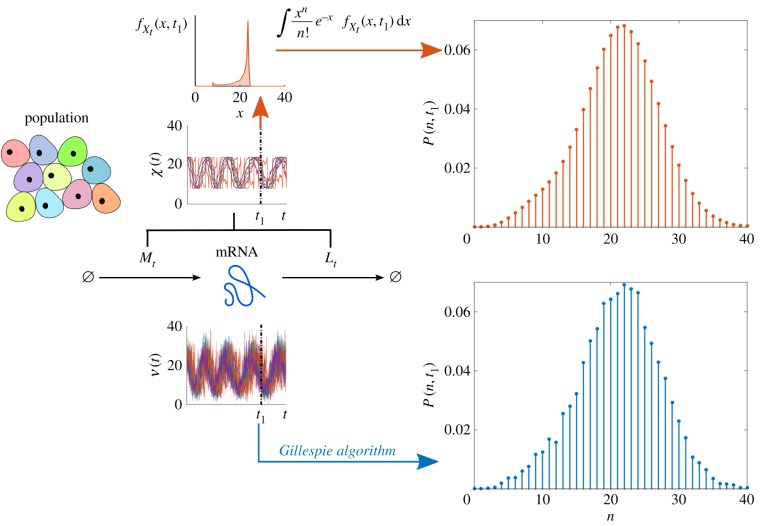
Figure 10.
Efficient sampling of the full distribution P(n, t) for transcription with upstream cellular drives. We consider upstream drives governed by the Kuramoto promoter model (3.4) for C = 10 000 coupled oscillatory cells. Sample paths of N are simulated directly with the Gillespie algorithm to approximate P(n, t) at time t1 (bottom, blue). Alternatively, sample paths of X are used to estimate 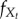 , which is then mixed by performing the numerical integration (2.14) to obtain P(n, t) (top, red). The latter sampling through X is more regular and far less costly: CPU time via N is ≈36000 s, whereas CPU time via X is ≈0.1 s. (Online version in colour.)
, which is then mixed by performing the numerical integration (2.14) to obtain P(n, t) (top, red). The latter sampling through X is more regular and far less costly: CPU time via N is ≈36000 s, whereas CPU time via X is ≈0.1 s. (Online version in colour.)