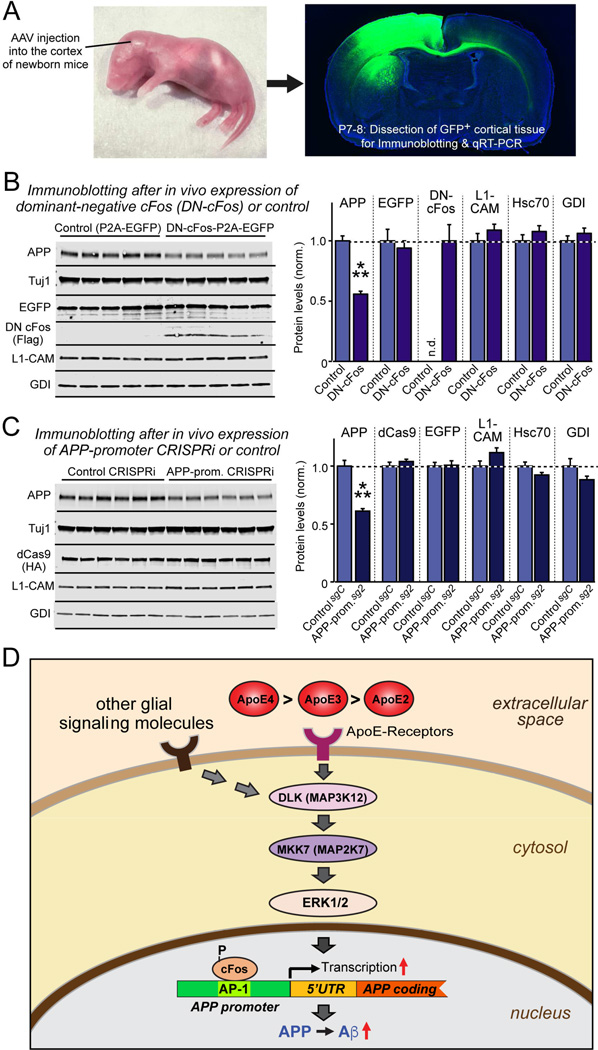
Figure 7. cFos-dependent signaling pathway regulates mouse App gene transcription in vivo.
(A) Experimental design. AAVs (encoding EGFP alone or EGFP and dominant-negative cFos (DN-cFos); and EGFP plus dCAS9 with either a control guide RNA or a guide RNA directed to the App promoter AP-1 binding site) were stereotactically injected into the cortex of anesthetized newborn mice (left), and cortex expressing EGFP was analyzed at P7-P8 (right).
(B & C) Suppression of cFos-signaling using DN-cFos (A; n = 5 mice for test and control) or CRISPRi of the AP-1 binding sequence of the App promoter (B; n = 6 mice for test and control) selectively decreases APP expression in vivo (for mRNA measurements, see Fig. S7). Data are means ± SEM (n.d., not detectable); statistical significance was evaluated with Student’s t-test (***, p<0.001).
(D) Schematic of the ApoE-signaling pathway that controls APP transcription and Aβ production via activation of the DLK MAP-kinase cascade. See text for details. ApoE is proposed to increase AD risk by causing an incremental chronic increase in APP abundance and Aβ secretion, with ApoE4 being more, and ApoE2 being less efficacious than ApoE3 in a parallel to their effects on AD risk.