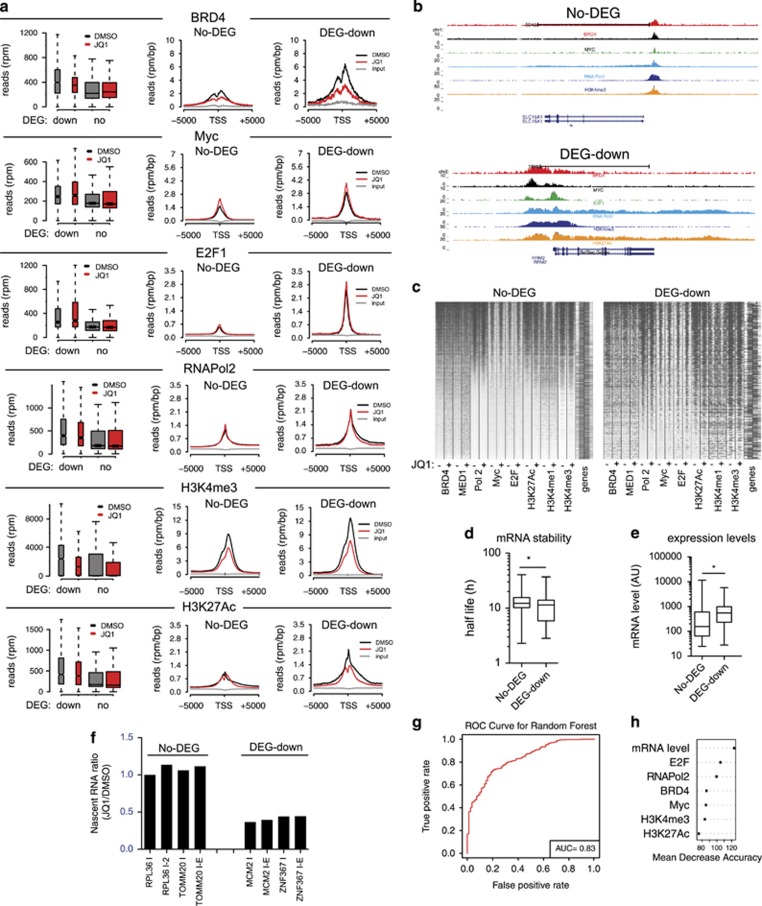
Figure 4.
Analysis of promoter occupancy in JQ1-sensitive genes. RAJI cells were treated with 100 nm JQ1 or vehicle (DMSO) for 24 h. Genes were subsetted based on their differential expression following JQ1 treatment in DEG-down (genes downregulated) or No-DEG (genes not affected by JQ1). (a) The enrichment of RNAPol2, BRD4, Myc, E2F and selected chromatin marks is reported as a box plot (left) and as cumulative reads distribution (center and right panels) calculated around the TSS. (b) Genome browser views of representative DEG-down or No-DEG genes. (c) Ranked heatmap showing the distribution of transcription factors, RNAPol2 and chromatin marks on the promoter of either DEG-down or No-DEG genes. For each factor the signal relative to mock-treated cells (−) and JQ1-treated cells (+) is shown. (d) Box plot showing the analysis of mRNA stability based on Schwanhäusser et al.63 *P-value <0.05 (Student's t-test). (e) Box plot showing relative expression levels of either DEG-down or No-DEG genes, based on μArray analysis. *P-value <0.05 (Student's t-test). (f) Nascent mRNA analysis of selected DEG-down or No-DEG genes performed by 4-sU labeling. Data are reported as normalized to the values determined for mock-treated cells (g) Receiver operating characteristic (ROC) curve associated to the Random Forest classifier. Area under the curve (AUC) shows that the response to JQ1 treatment (downregulation of a gene) could be predicted using the expression levels and the transcription factors and chromatin marks enrichments on the TSSs. (h) Predictive power of the features used in g. Features were ranked based on their variable importance in the prediction of JQ1 response. All the features contributed significantly to the prediction, with gene expression level, E2F and RNAPol2 enrichments being among the most predictive.