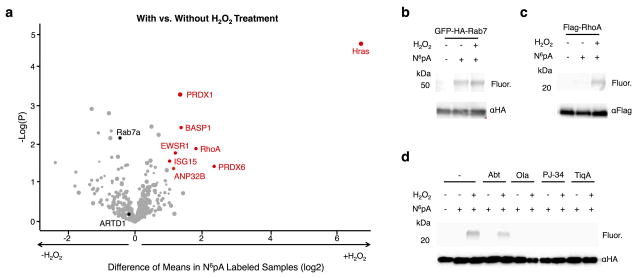
Figure 3.
Summary and validation of N6pA-labeled proteins. a) The scatter plot of N6pA-labeled proteins in both H2O2-treated and untreated cells. Protein hits were annotated by Maxquant analysis of recovered peptides. The x-axis is the difference of means between the H2O2-treated and untreated samples and the y-axis is the log of the probability of that difference determined by the t-test. The minimum values for a valid protein are p<0.05 and a difference of means of 2. Notable proteins are highlighted in black. b) HEK293T cells were transfected with GFP-HA-Rab7, labeled with 50 μM N6pA for 8 h and treated with 4 mM H2O2 for 1 h. GFP-HA-Rab7 was immunoprecipitated from cell lysates with antibody-conjugated beads, reacted with az-rho and analyzed in-gel fluorescence scanning. Western blots are included for protein loading control. c) N6pA labeling of FLAG-RhoA. d) N6pA labeling of HA-Hras after treatment with ADP-ribosylation inhibitors, ABT-888 (Abt, 10 μM), olaparib (Ola, 10 μM), PJ-34 (100 μM), and Tiq-A (20 μM), 2 h prior to N6pA labeling and H2O2-treatment. Full gels are located in Supplementary figure 9.