Figure 1.
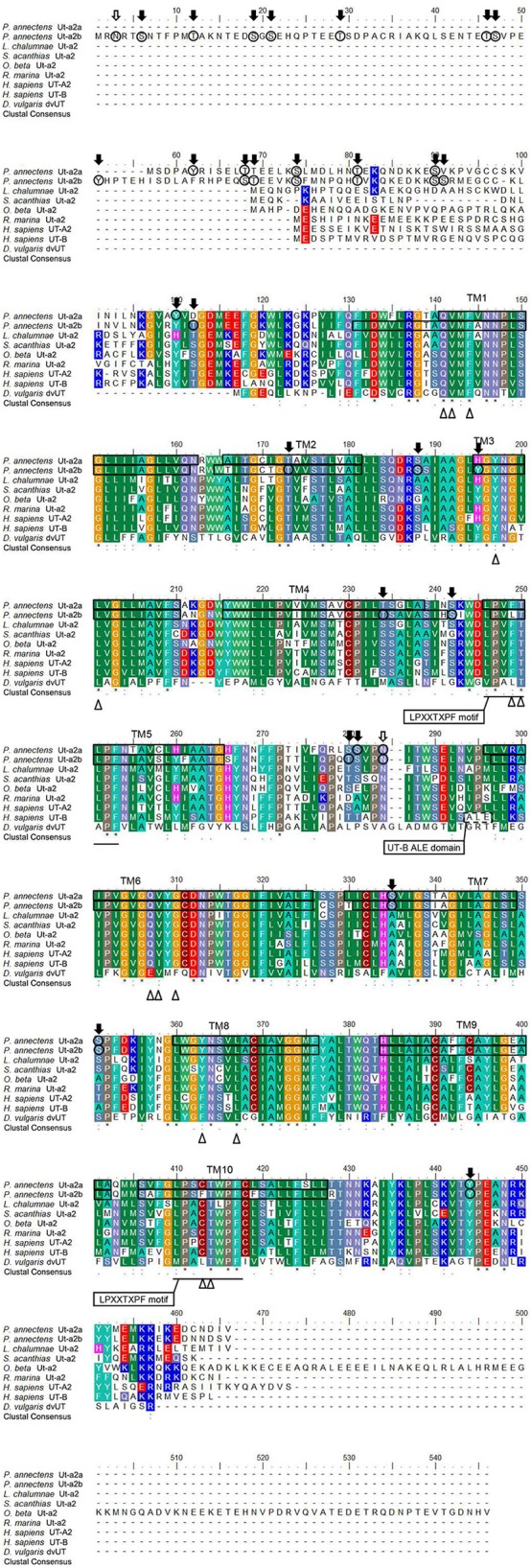
A multiple amino acid alignment of the isoforms of urea transporter (Ut-a2a and Ut-a2b) from Protopterus annectens with Latimeria chalumnae Ut-a2 (XP_006007026.1), Squalus acanthias Ut-a2 (AAF66072.1), Opsanus beta Ut-a2 (AAD53268.2), Rhinella marina Ut-a2 (BAE16706.1), Homo sapiens UT-A2 (CAA65657.1) and UT-B1 (CAB60834.1), and Desulfovibrio vulgaris Ut (Q72CX3). Identical amino acid residues are indicated by asterisks, strongly similar amino acids are indicated by colons and weakly similar amino acids are indicated by periods. The conserved LPXXTXPF motifs are underlined. The potential urea binding sites are indicated by open triangles. The dotted box denotes the UT-B signature ALE domain. Potential N-glycosylation and phosphorylation sites are indicated by open and shaded arrows, respectively. The predicted transmembrane domains (TM1-10) of Ut-a2a and Ut-a2b of P. annectens are indicated by open boxes and were predicted using MEMSATS and MEMSAT-SVA provided by PSIPRED protein structure prediction server.
