ABSTRACT
The aim of this study was to elucidate the antibacterial mechanism of 405 ± 5-nm light-emitting diode (LED) illumination against Salmonella at 4°C in phosphate-buffered saline (PBS) by determining endogenous coproporphyrin content, DNA oxidation, damage to membrane function, and morphological change. Gene expression levels, including of oxyR, recA, rpoS, sodA, and soxR, were also examined to understand the response of Salmonella to LED illumination. The results showed that Salmonella strains responded differently to LED illumination, revealing that S. enterica serovar Enteritidis (ATCC 13076) and S. enterica subsp. enterica serovar Saintpaul (ATCC 9712) were more susceptible and resistant, respectively, than the 16 other strains tested. There was no difference in the amounts of endogenous coproporphyrin in the two strains. Compared with that in nonilluminated cells, the DNA oxidation levels in illuminated cells increased. In illuminated cells, we observed a loss of efflux pump activity, damage to the glucose uptake system, and changes in membrane potential and integrity. Transmission electron microscopy revealed a disorganization of chromosomes and ribosomes due to LED illumination. The levels of the five genes measured in the nonilluminated and illuminated S. Saintpaul cells were upregulated in PBS at a set temperature of 4°C, indicating that increased gene expression levels might be due to a temperature shift and nutrient deficiency rather than to LED illumination. In contrast, only oxyR in S. Enteritidis cells was upregulated. Thus, different sensitivities of the two strains to LED illumination were attributed to differences in gene regulation.
IMPORTANCE Bacterial inactivation using visible light has recently received attention as a safe and environmentally friendly technology, in contrast with UV light, which has detrimental effects on human health and the environment. This study was designed to understand how 405 ± 5-nm light-emitting diode (LED) illumination kills Salmonella strains at refrigeration temperature. The data clearly demonstrated that the effectiveness of LED illumination on Salmonella strains depended highly on the serotype and strain. Our findings also revealed that its antibacterial mechanism was mainly attributed to DNA oxidation and a loss of membrane functions rather than membrane lipid peroxidation, which has been proposed by other researchers who studied the antibacterial effect of LED illumination by adding exogenous photosensitizers, such as chlorophyllin and hypericin. Therefore, this study suggests that the detailed antibacterial mechanisms of 405-nm LED illumination without additional photosensitizers may differ from that by exogenous photosensitizers. Furthermore, a change in stress-related gene regulation may alter the susceptibility of Salmonella cells to LED illumination at refrigeration temperature. Thus, our study provides new insights into the antibacterial mechanism of 405 ± 5-nm LED illumination on Salmonella cells.
KEYWORDS: 405-nm light-emitting diode, DNA oxidation, Salmonella, antibacterial mechanism, gene expression, membrane functions
INTRODUCTION
Salmonella is a major causative agent of foodborne illness worldwide, resulting in approximately 80.3 million cases of infection and 155,000 deaths each year (1). According to the Foodborne Disease Active Surveillance Network (FoodNet) in the United States, among a total of 19,542 cases of infection, 7,452 infections, 2,141 hospitalizations, and 30 deaths were caused by Salmonella in 2014 (2). In Singapore, Salmonella also has been identified as the top contributor to foodborne illness, accounting for more than half of the reported cases from 2001 to 2010 (3). To minimize the risk of salmonellosis, storing perishable and ready-to-eat foods in the refrigerator is a common practice. However, Salmonella cells, having a cold shock protein (CspH), are able to survive at temperatures lower than 5°C (4). Moreover, refrigerators in retail shops are often held at temperatures higher than 5°C (5), enabling potential growth of Salmonella and thus increasing the risk of salmonellosis. For these reasons, a secondary hurdle in combination with low temperature should be employed to more effectively control Salmonella in foods during storage.
A light-emitting diode (LED) with visible-light wavelengths has recently received increased attention as an emerging food preservation technology due to its antibacterial effect, described as photodynamic inactivation (PDI). Results from previous studies showed that blue LEDs have antibacterial efficacy against pathogenic bacteria in buffered solutions and food matrices with or without the addition of exogenous photosensitizers, such as chlorophyll and porphyrin (6–8). In our previous studies, we demonstrated the antibacterial effects of 405- and 460-nm LEDs against Bacillus cereus, Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar Typhimurium, Shigella sonnei, and Staphylococcus aureus in buffered solutions and bacterial growth media under chilling conditions without the aid of a photosensitizer (9–12).
The proposed mechanism of PDI requires three components: visible light in the wavelength range of 400 to 430 nm, photosensitizers, and oxygen (6). Once bacterial cells are exposed to light, the photosensitizer, such as intracellular porphyrin compounds, absorbs light energy and is then excited, resulting in the production of reactive oxygen species (ROS) by transferring energy to oxygen. ROS, such as hydrogen peroxide, superoxide, and singlet oxygen, can result in cytotoxicity by reacting with DNA, RNA, proteins, and lipids, eventually causing bacterial death (6, 11). However, the detailed mechanism of the antimicrobial activity of 405 ± 5-nm LED illumination in the absence of an exogenous photosensitizer has not been described. We hypothesized that the mode of bacterial inactivation of LED illumination without additional photosensitizers may differ from that by exogenous photosensitizers, since genomic DNA and membrane proteins of Salmonella cells may be major targets of ROS generated by 405 ± 5-nm LED illumination due to the cellular localization of an intracellular endogenous photosensitizer.
It is crucial to understand the response of Salmonella to oxidative stress by ROS generated from LED illumination for elucidating its antibacterial mechanism. It is known that the resistance of Salmonella to oxidative stress is enhanced by the induction of regulators, such as OxyR, SodA, SoxRS, and RpoS (13). The gene encoding the OxyR regulator, oxyR, is activated via the formation of disulfide bonds under hydrogen peroxide stress (13). The oxidized OxyR also activates the expression of katG (encoding catalase hydroperoxidase I), dps (encoding nonspecific DNA-binding proteins), gorA (encoding glutathione reductase), and oxyS (encoding small regulatory RNA) involved in the bacterial resistance to oxidative stress (13–15). Bacteria possess at least two superoxide dismutases (SODs), Mn-SOD encoded by sodA and Fe-SOD encoded by sodB, which catalyze the conversion of superoxide anions into hydrogen peroxide and oxygen. The SoxRS regulator, encoded by two regulatory genes, soxR and soxS, regulates SODs under high levels of superoxide stress (13–15). The rpoS-encoded σS subunit of RNA polymerase is another regulator that survives under oxidative stress, as well as under general stresses, including low temperature and starvation (16, 17). It is known that UV light and hydrogen peroxide (at low and high concentrations) induce recA as a global regulator of the SOS response (13); recA expression is associated with DNA topological change, DNA gyrase, and single-stranded DNA (13, 18).
Although the molecular response of Salmonella to general oxidative stress has been well documented (13), its response to LED illumination at a molecular level is unknown. Furthermore, studies to determine if differences in gene regulation influence bacterial susceptibility to LED illumination have not been reported. Thus, the objective of this study was to elucidate the detailed antibacterial mechanism of 405 ± 5-nm LED illumination on Salmonella by determining changes in endogenous coproporphyrin content, in DNA oxidation, in the amount of damage to membrane functions, in morphology, and in regulatory gene expression levels during LED illumination at refrigeration temperature.
RESULTS
Temperature change caused by LED illumination.
Changes in temperatures of tryptone soya agar (TSA) and phosphate-buffered saline (PBS) during 405 ± 5-nm LED illumination were monitored over a 5-h period at 1-min intervals at a set temperature of 4°C. LED illumination increased temperatures by approximately 4.5°C on the surface of TSA and PBS within 1 h (data not shown). Thus, nonilluminated control cells were held at 9.5°C to eliminate the difference in temperatures under LED illumination and nonilluminated conditions.
Antibacterial efficacy of LED illumination against Salmonella.
Eighteen strains of Salmonella enterica plated onto the surface of TSA were illuminated at doses of 288 J/cm2 with a 405 ± 5 nm LED at a set temperature of 4°C to select for resistance. This experiment was performed in three independent trials with duplicate TSA plates in each trial (n = 6). Representative photographs of plates incubated at 37°C for 24 h are presented in Fig. 1. The results show that S. enterica subsp. enterica serovar Enteritidis ATCC 13076 and S. enterica subsp. enterica serovar Saintpaul ATCC 9712 were the most susceptible and resistant strains to LED illumination, respectively. These two strains were selected for further experiments.
FIG 1.
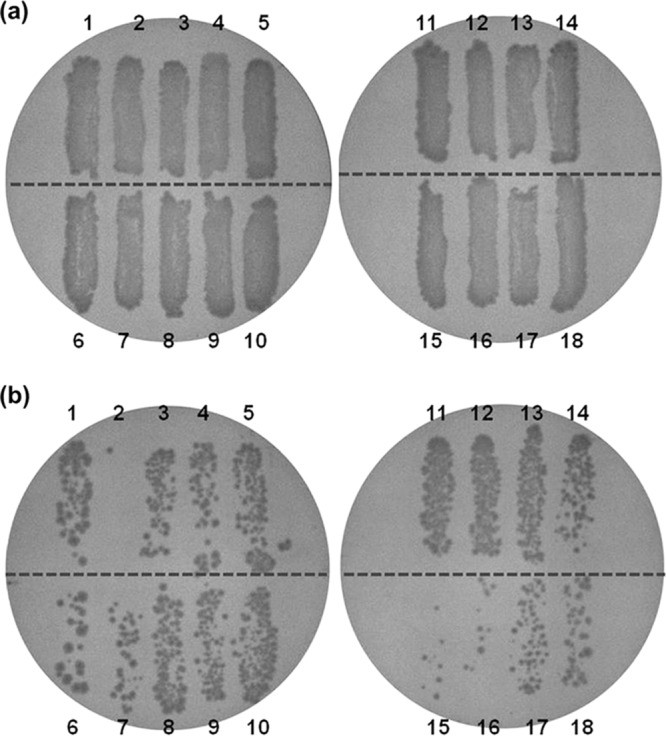
Antibacterial effects of 405 ± 5-nm LED illumination at 288 J/cm2 against 18 Salmonella strains at a set temperature of 4°C (actual temperature of 9.5°C) on TSA plates. (a) Nonilluminated Salmonella. (b) LED-illuminated Salmonella. Lane 1, S. Agona; lane 2, S. Enteritidis ATCC 13076; lane 3, S. Enteritidis 109; lane 4, S. Enteritidis 124; lane 5, S. Enteritidis 125; lane 6, S. Enteritidis 130; lane 7, S. Gaminara; lane 8, S. Heidelberg; lane 9, S. Montevideo; lane 10, S. Newport; lane 11, S. Poona; lane 12, S. Saintpaul; lane 13, S. Tennessee; lane 14, S. Typhimurium ATCC 14028; lane 15, S. Typhimurium ATCC 13311; lane 16, S. Typhimurium ATCC 25241; lane 17, S. Typhimurium ATCC 29269; lane 18, S. Typhimurium ATCC 51812.
Suspensions of S. Enteritidis and S. Saintpaul cells were illuminated with a 405 ± 5-nm LED at a set temperature of 4°C and at doses as high as 576 J/cm2 to quantitatively measure inactivation. Regardless of the strain, the numbers of nonilluminated control cells remained unchanged during storage at 9.5°C, whereas the populations of LED-illuminated S. Enteritidis and S. Saintpaul cells significantly (P ≤0.05) decreased when exposed to 144 J/cm2 (Fig. 2). The difference in the extents of inactivation between the two strains was obvious when cells were exposed to illumination at 288 J/cm2. Populations of S. Enteritidis and S. Saintpaul were reduced by 2.0 and 1.0 log CFU/ml at 288 J/cm2, respectively, and by 5.6 and 1.7 log CFU/ml at 576 J/cm2, respectively. Moreover, D values (J/cm2; i.e., dosages required to bring about 1-log reduction) for the first-order inactivation kinetics were calculated from the linear portion of the survival curve to compare the bacterial sensitivities to 405 ± 5-nm LED illumination (Table 1). The D value of S. Saintpaul cells was 2.3 times higher than that of S. Enteritidis cells, indicating that S. Enteritidis was more sensitive to 405 ± 5-nm LED illumination than was S. Saintpaul.
FIG 2.
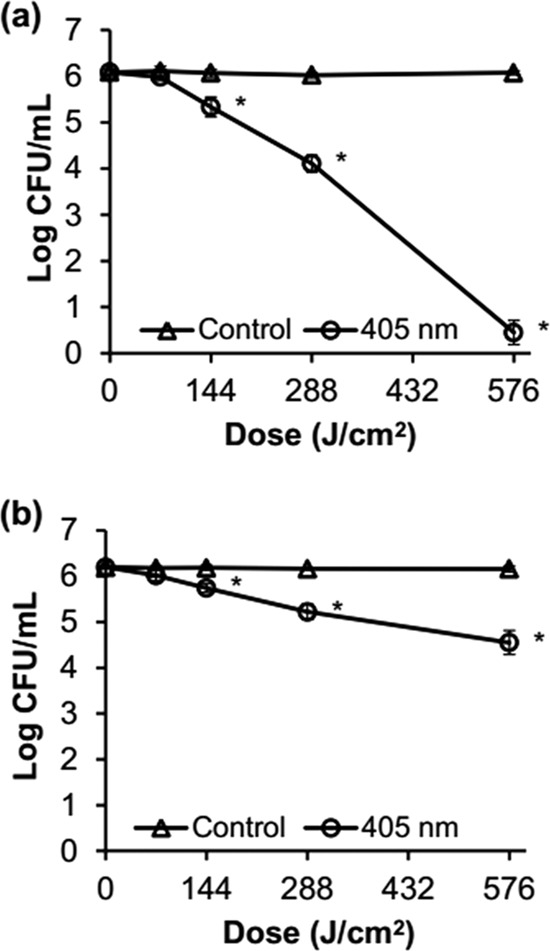
Survival curves for S. Enteritidis ATCC 13076 (a) and S. Saintpaul ATCC 9712 (b) during 405 ± 5-nm LED illumination at a set temperature of 4°C and doses as high as 576 J/cm2 in PBS. The detection limit was 1 CFU/ml. *, P ≤ 0.05 versus control cells.
TABLE 1.
Comparison of D values from S. Enteritidis ATCC 13076 and S. Saintpaul ATCC 9712 after 405 ± 5-nm LED illumination at a set temperature of 4°C
| Strain | D value (J/cm2)a | R2 |
|---|---|---|
| S. Enteritidis | 146.4 ± 12.4 | 0.97 ± 0.01 |
| S. Saintpaul | 338.8 ± 51.2 | 0.99 ± 0.01 |
D values for the two strains were significantly different from each other (n = 6; P ≤ 0.05).
Intracellular coproporphyrin contents.
The amounts of intracellular coproporphyrin compounds in stationary-phase S. Enteritidis and S. Saintpaul cells were quantified to better understand differences in inactivation patterns between the two strains. The total amounts of coproporphyrin in S. Enteritidis and S. Saintpaul were 61.1 and 46.5 ng/mg, respectively, indicating no significant (P >0.05) difference between the two strains (data not shown).
DNA oxidation.
Exposures of S. Enteritidis and S. Saintpaul cells to LED illumination at 576 J/cm2 resulted in 5.1 and 1.9 log reductions, respectively. Genomic DNA was extracted from illuminated cells to determine if ROS produced during LED illumination oxidized DNA, particularly guanine. Levels of 8-hydroxydeoxyguanosine (8-OHdG) in LED-illuminated S. Enteritidis and S. Saintpaul were 2.8 and 2.6 times higher (P ≤ 0.05), respectively, than those in nonilluminated cells, but there was no significant (P > 0.05) difference in the levels of 8-OHdG in the two strains (Fig. 3).
FIG 3.
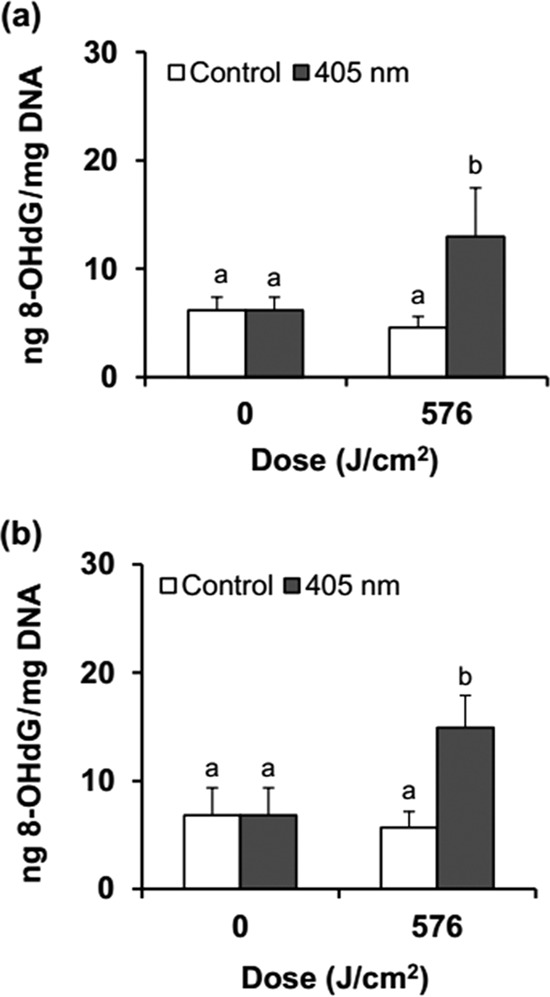
Changes in the amounts of 8-hydroxydeoxyguanosine (8-OHdG) in S. Enteritidis (a) and S. Saintpaul (b) by 405 ± 5-nm LED illumination at a set temperature of 4°C. Different letters (a, b) for each dose indicate significantly different (P ≤ 0.05) values.
Damage to membrane function.
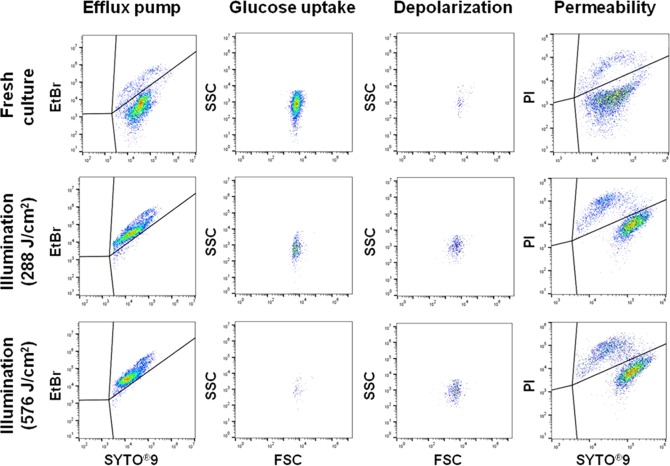
Damage to cellular membrane function was examined using flow cytometry with five probes. SYTO9, propidium iodide (PI), and ethidium bromide (EtBr) bind to nucleic acids, and green-fluorescing SYTO9 enters cells through intact as well as damaged membranes, while red-fluorescing PI penetrates only the damaged cytoplasmic membrane (10, 19). Red-fluorescing EtBr enters only nonpumping cells, since intact cells can actively release EtBr through efflux pumping, a nonspecific proton transport system (19). When SYTO9 coexists with either PI or EtBr in cells, the intensity of SYTO9 is reduced. Green-fluorescing bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] binds to intracellular proteins by entering cells through depolarized or damaged cytoplasmic membranes (19). A fluorescent glucose analogue, 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy–d-glucose (2-NBDG), monitors glucose uptake via the glucose-specific phosphoenolpyruvate-phosphotransferase system (PEP-PTS) in intact cells as an indicator of cellular viability (19).
The percentages of membrane function lost were calculated based on flow cytometry plots (Fig. 4) and are presented in Fig. 5. Except for glucose uptake activity by S. Enteritidis, the results showed that no significant (P >0.05) changes in membrane functions occurred in nonilluminated control cells during storage at 9.5°C. On the other hand, the percentages of efflux pump activity lost increased up to 98% for S. Enteritidis cells and to 99% for S. Saintpaul cells dosed with 288 J/cm2 (Fig. 5a). The flow cytometry plots (Fig. 6) also clearly show an increase in the intensity of red-fluorescing EtBr in illuminated S. Enteritidis cells compared to that of freshly cultured cells. For glucose uptake activity, loss percentages were 76% for S. Enteritidis and 67% for S. Saintpaul at 288 J/cm2, followed by increases up to 99% for both strains after exposures to 576 J/cm2 (Fig. 5b). The percentages of membrane depolarization and permeability of both Salmonella serotypes were significantly (P ≤0.05) increased by LED illumination. However, the difference between nonilluminated and illuminated cells was only 10% or less (Fig. 5c and d). Among the membrane functions analyzed in this study, a significant (P ≤ 0.05) difference was observed in the percentages of membrane permeability between S. Enteritidis and S. Saintpaul after LED illumination, showing that the permeability of S. Enteritidis was 8.6% higher than that of S. Saintpaul.
FIG 4.
Flow cytometry plots of S. Enteritidis cells stained with SYTO9/EtBr, SYTO9/PI, DiBAC4(3), or 2-NBDG after 405 ± 5-nm LED illumination at a set temperature of 4°C.
FIG 5.
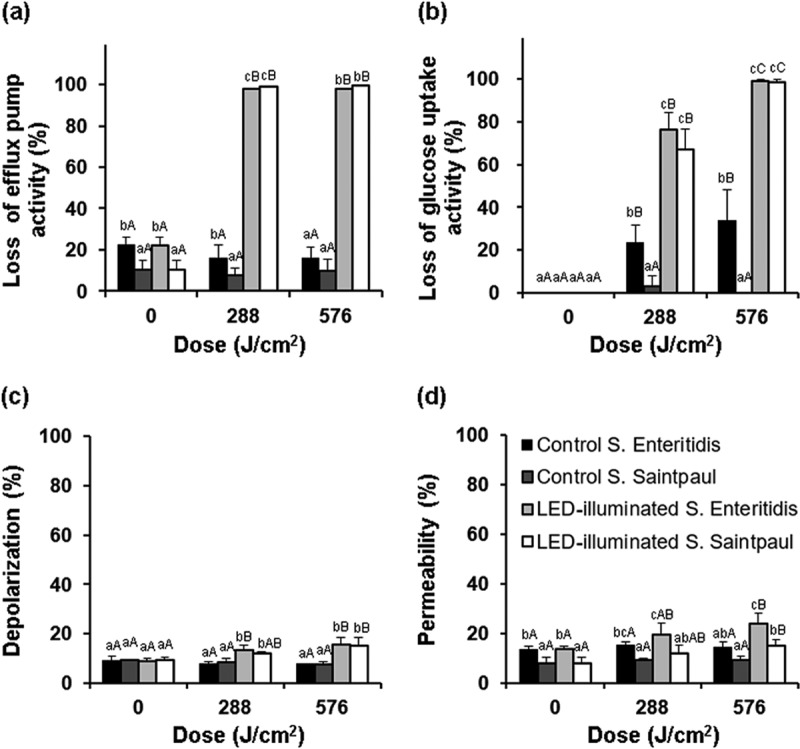
Changes in the membrane functions of S. Enteritidis and S. Saintpaul during 405 ± 5-nm LED illumination at a set temperature of 4°C. (a) Loss of efflux pump activity measured with SYTO9/EtBr. (b) Loss of glucose uptake activity measured with 2-NBDG. (c) Loss of membrane potential measured with DiBAC4(3). (d) Loss of membrane integrity measured with SYTO9/PI. Uppercase letters (A, B, C) for the same bar and lowercase letters (a, b, c) for the same exposure dose indicate significant differences (P ≤ 0.05).
FIG 6.
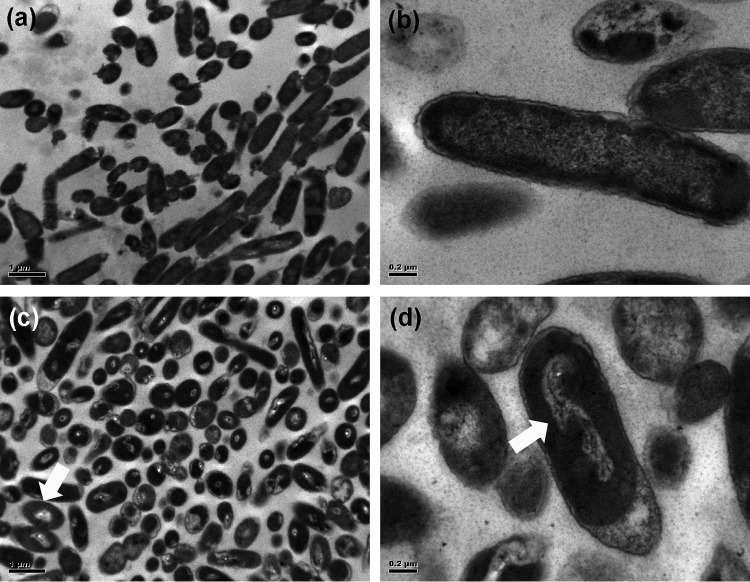
Micrographs of nonilluminated and LED-illuminated S. Enteritidis cells exposed to 720 J/cm2 at a set temperature of 4°C. (a) Nonilluminated control cells at ×10,000 magnification. (b) Nonilluminated control cells at ×40,000 magnification. (c) LED-illuminated cells at ×10,000 magnification. (d) LED-illuminated cells at ×40,000 magnification. The morphological changes in illuminated cells are highlighted by arrows.
Transmission electron microscopy.
To obtain concrete evidence that 405 ± 5-nm LED illumination causes changes in external and internal cell structures, illuminated and nonilluminated cells were examined by transmission electron microscopy (TEM). S. Enteritidis was chosen as a model strain for this analysis. TEM images revealed that the cytoplasm of nonilluminated cells was evenly distributed without an aggregation of cellular components (Fig. 6a and b), while LED-illuminated cells had altered appearances with disorganized genomic areas (Fig. 6c and d). This indicates that LED illumination primarily induces intracellular damage rather than outer membrane damage, although some of the nonilluminated and illuminated cells showed nondistinctive outer membranes with a collapsed appearance.
Changes in gene expression levels caused by LED illumination.
The levels of relative gene expression in nonilluminated and illuminated S. Enteritidis and S. Saintpaul cells are presented in Fig. 7. The results showed that only oxyR was upregulated in nonilluminated and illuminated S. Enteritidis cells (Fig. 7a), whereas transcription levels of all the genes tested (oxyR, recA, rpoS, sodA, and soxR) were upregulated in nonilluminated and illuminated S. Saintpaul cells (Fig. 7b). Among the five genes, the transcription levels of oxyR in both strains were significantly (P ≤0.05) different in nonilluminated cells compared with those in illuminated cells. However, the transcription level of oxyR was 1.5 times higher in illuminated S. Enteritidis cells and 1.4 times lower in illuminated S. Saintpaul cells compared with the levels in nonilluminated cells.
FIG 7.
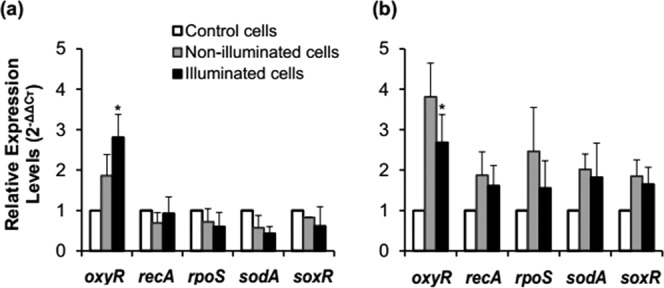
Relative expression levels of oxyR, recA, rpoS, sodA, and soxR in nonilluminated and LED-illuminated cells of S. Enteritidis (a) and S. Saintpaul (b) at a set temperature of 4°C at 72 J/cm2. *, P ≤ 0.05 versus nonilluminated cells.
DISCUSSION
Although the results of our previous studies demonstrated the antibacterial effect of a 405 ± 5-nm LED on various foodborne pathogens and the damage to bacterial membranes (10, 11), the detailed antibacterial mechanism of LED illumination on a given bacterial species had not been studied. Thus, this study was designed to assess the efficacy of LED illumination against Salmonella, to elucidate its antibacterial mechanism at membrane and gene levels, and to identify the factors influencing the differences in sensitivities to LED illumination.
The antibacterial effect of 405 ± 5-nm LED illumination was evaluated at a set temperature of 4°C. The effectiveness of LED illumination is known to be enhanced at chilling temperatures rather than at room temperature (9). Our results show that the antibacterial efficacy of 405 ± 5-nm LED illumination varies among Salmonella strains grown on TSA plates (Fig. 1). Of the 18 strains tested, S. Enteritidis ATCC 13076 and S. Saintpaul ATCC 9712 were found to be the most sensitive and resistant strains, respectively. Four S. Typhimurium strains and five S. Enteritidis strains also exhibited differing sensitivities to LED illumination. The difference in the sensitivity to LED illumination between S. Enteritidis ATCC 13076 and S. Saintpaul ATCC 9712 was more obvious in PBS than on TSA (Fig. 2). This indicates that efficacy of LED illumination may depend on serotype and strain within the same serotype. In contrast, Endarko et al. (20) reported similar sensitivities to 405-nm LED illumination among Listeria ivanovii, L. monocytogenes, and Listeria seeligeri in PBS, while the sensitivities of S. Enteritidis, S. sonnei, L. monocytogenes, and E. coli differed. Results from another study showed that treatment with a 405-nm LED in PBS caused 5-log reductions of L. monocytogenes, S. sonnei, and E. coli O157:H7 at different doses (21). Similar to our results, variations in sensitivities to UV irradiation have also been reported. For example, Gayán et al. (22) similarly showed differing sensitivities to UV light among five strains of Salmonella, revealing that S. Enteritidis ATCC 13076 and S. Typhimurium STCC 878 were the most sensitive and resistant, respectively.
Bacterial inactivation by LED illumination without any exogenous photosensitizer is theoretically due to the intracellular ROS generated by excited endogenous porphyrins naturally present in the cells (6, 11). These ROS could nonspecifically damage cellular components, such as DNA, proteins, and lipids, eventually causing cell death. It is known that protoporphyrin IX, an endogenous photosensitizer, localizes to the cytoplasm and is catalyzed by ferrochelatase, one of the cytoplasmic enzymes involved in incorporating iron in the final step of heme synthesis (23). Thus, we hypothesized that differing photodynamic inactivation rates in S. Enteritidis and S. Saintpaul might be attributed to different levels of endogenous porphyrin compounds. Since coproporphyrin is a precursor of protoporphyrin IX (14), we quantified the total amounts of coproporphyrin in the two Salmonella strains by high-pressure liquid chromatography (HPLC) in this study. Our data showed that there was no significant (P >0.05) difference in the amounts present in the two strains, indicating that coproporphyrin content is not a contributing factor to the differences in sensitivity to LED illumination (data not shown). Similar results reported by Kumar et al. (12) showed that S. Typhimurium produced a larger amount of coproporphyrin than did E. coli O157:H7, but the inactivation rate of S. Typhimurium was lower than that of E. coli O157:H7 during 405-nm LED illumination. In addition, B. cereus, with a larger amount of coproporphyrin than S. aureus, was less susceptible to 405-nm LED illumination than was S. aureus. These data suggest that there may not be a relationship between levels of coproporphyrin and inactivation rates among bacteria and indicate that other factors, e.g., the extent of cellular damage, are more likely to reflect the differences in cell susceptibilities to LED illumination.
Since cellular DNA is thought to be the major target of ROS generated by LED illumination, the oxidized derivative of 8-OHdG was first analyzed to determine if LED illumination causes DNA oxidation. Our results show that levels of 8-OHdG in S. Enteritidis and S. Saintpaul cells increased significantly upon LED illumination, confirming that exposure to a 405 ± 5-nm LED oxidizes genomic DNA by generating intracellular ROS (Fig. 3). However, there was no difference in the degrees of DNA oxidation between the two strains. Similar to our results, other studies have reported DNA oxidation in S. Typhimurium by 365-nm LED illumination (23) and in E. coli by illumination at 407 nm in the presence of tetra-meso (N-methyl pyridyl) porphine (TMPyP), an exogenous photosensitizer (24).
The loss of cellular membrane functions caused by LED illumination was measured using flow cytometry. Our results show that LED illumination completely inactivates efflux pumps (Fig. 4 and 5a). Efflux pumps, which transport proteins, localize to and are embedded in the cytoplasmic membrane and recognize unwanted agents, such as antibiotics and EtBr in the environment and cytotoxic products produced during metabolism (25). To move noxious agents from the inside out, the efflux pump utilizes energy sources through proton motive force (PMF) or ATP synthesis (25). Thus, one possibility for rationalizing the loss of efflux pump activity is that intracellular ATP is depleted due to the inhibition of ATPase by ROS (19, 26). This is also associated with the abortion of DNA replication, resulting in no bacterial growth (26). A previous study conducted by Berney, Weilenmann, and Egli (19) demonstrated that UV-A light rapidly diminished ATP and simultaneously inactivated efflux pump activity. Another possibility is that ROS are generated by LED illumination from cytochromes containing a heme prosthetic group in the cytoplasmic membranes (27), which could affect nearby efflux pumps. Nitzan and Ashkenazi (28) reported that 405-nm light illumination in the presence of TMPyP not only damages the sodium-potassium pump but also inactivates enzymes, such as NADH dehydrogenase and lactate dehydrogenase. Based on these observations, it is speculated that intracellular ROS produced in the response to LED illumination might attack proteins associated with the function of the efflux pump.
Our results show that the glucose uptake system, i.e., glucose-specific PEP-PTS, in Salmonella was also extensively damaged by LED illumination, regardless of the strain. It is known that this system catalyzes a group translocation process to phosphorylate glucose through a phosphoryl transfer process and monitors metabolism in response to the availability of consumable glucose (29, 30). The damage to PEP-PTS caused by LED illumination might be due, in part, to the inactivation of enzymes associated with PEP-PTS and ATP synthesis utilized for phosphorylation. Surprisingly, nonilluminated S. Enteritidis, but not S. Saintpaul, also had a slight loss in glucose uptake activity (approximately 34%) at refrigeration temperature. This may be because S. Enteritidis was more sensitive to cold and starvation conditions than S. Saintpaul.
A previous study testing the effects of UV-A and sunlight on E. coli also reported damage to the efflux pump and glucose uptake system (19). Efflux pump activity initially ceased, and then membrane potential and glucose uptake were gradually lost, and considerable membrane permeability occurred. Our results show only a 6 to 10% higher level in cellular depolarization and permeability after LED illumination compared with levels in freshly cultured and nonilluminated control cells. Our previous studies also showed that only some of the illuminated S. Typhimurium cells lost membrane integrity under the same illumination conditions (11). Similar to our observations, Caminos et al. (31) reported no change in the membrane integrity of E. coli after PDI with the aid of an exogenous photosensitizer, although membrane functions could have been lost. The study by Alves et al. (32) demonstrated that large amounts of exogenous photosensitizers might be required to disrupt the outer cell membrane by PDI. Thus, it is speculated that the small amounts of endogenous porphyrins in Salmonella cells do not generate sufficient ROS to disrupt cell membrane integrity during LED illumination. However, ROS might be present in amounts sufficient to oxidize DNA and affect efflux pump activity and glucose uptake systems due to their proximity to endogenous porphyrins. This possibility appears to be supported by the results obtained by Kumar et al. (12), who reported 9 to 25 times lower concentrations of coproporphyrin compounds in S. Typhimurium cells than in Gram-positive bacteria. Moreover, the slight increases in membrane depolarization and permeability caused by LED illumination might be due to losses in efflux pump activity and other membrane proteins (19, 33).
To better understand the antibacterial mechanism resulting from LED illumination, we used TEM to examine the morphological damage of illuminated cells. We observed visual changes in nucleoid and ribosome areas, but we did not find noticeable damage in the cell envelope (Fig. 6). These results correlate with the observations on DNA oxidation and low levels of membrane depolarization and permeability. These findings clearly indicate that photodynamic inactivation of LED illumination without the addition of photosensitizers may not be due to membrane disruption.
We investigated the transcription levels of oxyR, recA, rpoS, sodA, and soxR in 405 ± 5-nm LED-illuminated S. Enteritidis (a sensitive strain) and S. Saintpaul (a resistant strain) cells to determine if these genes contribute to differences in sensitivity. Our results show that nonilluminated and LED-illuminated S. Saintpaul cells in PBS upregulated all five test genes, while nonilluminated and illuminated S. Enteritidis cells upregulated only oxyR (Fig. 7). This suggests that gene expression levels might be altered by exposure to refrigeration temperature, by starving the cells, or by both conditions rather than by LED illumination. However, upregulation under these conditions could make S. Saintpaul more resistant than S. Enteritidis to LED illumination. Similar to our findings, Smirnova, Zakirova, and Oktyabrskii (34) reported that the response of E. coli to a shift in temperature from 37 to 20°C within 10 min resembled oxidative stress responses, resulting in an upregulation of sodA, but no change in katG that is controlled by oxyR. Results from another study showed that the levels of oxyR expression in four Vibrio vulnificus strains were elevated by cold shock at 4°C, whereas transcription levels of katG were slightly changed in all strains but at different exposure times (35). In our study, rpoS and oxyR were upregulated in S. Saintpaul in response to chilling and starvation conditions. This may be a response of rpoS to the positive transcription regulation of oxyR for adapting to environmental changes (36). Merrikh et al. (37) reported that a number of antioxidant enzymes, e.g., catalase and SOD, are regulated by RpoS under oxidative stress. rpoS mutants of a S. Typhimurium strain were reported to be more sensitive than a wild-type strain to UV light (38). Inducing the RpoS regulator makes bacterial cells resistant to various stresses and starvation (17). Thus, the results obtained in our study indicate that different sensitivities of S. Enteritidis and S. Saintpaul to LED illumination might be due to the regulation of stress-related genes under refrigeration and starvation conditions.
Conclusions.
This comprehensive study elucidates the detailed antibacterial mechanism of 405 ± 5-nm LED illumination at refrigeration temperature against Salmonella cells and identifies factors influencing the sensitivity to LED illumination. Our results show that the differences in sensitivities of cells are strain and serotype dependent. The findings confirm that genomic DNA oxidation and the loss of membrane functions, preferentially in efflux pump and glucose uptake activities, are caused by LED illumination, but there is only slight damage to membrane potentials and integrity. As with findings on DNA oxidation, TEM images clearly show that genomes are one of the major targets of LED illumination. This indicates that the antibacterial mechanism of 405 ± 5-nm LED illumination may be due to DNA oxidation and a loss of membrane functions. Furthermore, all five of the regulatory genes of the LED-resistant S. Saintpaul were highly upregulated in response to low temperature and starvation rather than to illumination, suggesting that these changes make it more resistant than S. Enteritidis to LED illumination. These observations help us to better understand the antibacterial mechanism of 405 ± 5-nm LED illumination on Salmonella in the absence of exogenous photosensitizers.
MATERIALS AND METHODS
Bacterial culture conditions.
Eighteen Salmonella enterica strains were used in this study. S. Agona BAA-707 (alfalfa sprouts), S. Enteritidis ATCC 13076 (CDC), S. Gaminara BAA-711 (orange juice), S. Heidelberg ATCC 8326, S. Montevideo BAA-710 (clinical isolate from a tomato outbreak), S. Newport ATCC 6962 (food poisoning fatality, England), S. Poona BAA-1673 (iguana), S. Saintpaul ATCC 9712 (cystitis, Panama), S. Tennessee ATCC 10722, S. Typhimurium ATCC 14028 (poultry), S. Typhimurium ATCC 13311 (human feces), S. Typhimurium ATCC 25241 (methionine auxotroph derived from strain LT-2), S. Typhimurium ATCC 29269, and S. Typhimurium ATCC 51812 (human blood) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). S. Enteritidis 109 (phage type group D1; Peter Holt), S. Enteritidis 124 (phage type 8; Maryland Department of Health and Mental Hygiene, MD, USA), S. Enteritidis 125 (phage type 13A; U.S. Department of Agriculture, WA, USA), and S. Enteritidis 130 (phage type 2; CDC, NY, USA) were obtained from Kun-Ho Seo at Konkuk University, Republic of Korea. All Salmonella strains were stored at −70°C in Cryoinstant vials with porous beads (DeltaLab, Barcelona, Spain). Cells were activated in 10 ml of sterile tryptone soya broth (TSB; Oxoid, Basingstoke, UK) for 18 to 24 h at 37°C. After two consecutive transfers at 18- to 24-h intervals at 37°C, 1 ml of stationary-phase culture was centrifuged at 6,000 × g for 10 min at 4°C and washed twice with 1 ml of sterilized phosphate-buffered saline (PBS; Vivantis Technologies Sdn Bhd, Malaysia). The cells in the resultant pellet were resuspended in 1 ml of PBS and serially diluted to initial populations of approximately 106, 108, or 109 CFU/ml for LED illumination.
Light-emitting diode illumination system.
High-intensity 405 ± 5-nm LEDs (8 by 8 mm) were purchased from Shenzhen Getian Opto-Electronics Co., Ltd. (Shenzhen, Guangdong, China). The LED illumination system is described elsewhere (9, 10). Briefly, LEDs were attached to a heat sink and a fan to minimize heat transfer to the cell suspension. To prevent the penetration of external light during illumination, an acrylonitrile butadiene styrene (ABS) housing was constructed for each LED system. The suspension (10 ml and 1 cm in depth) in a sterile petri dish (60 mm diameter) or after streaking on TSA (1 cm depth; Oxoid) plates (90 mm diameter) was placed directly below the LED bulbs at a distance of 4.5 cm to illuminate the entire surface of the inoculum (Fig. 8). The surface temperatures of TSA and PBS were monitored with a Fluke 5.4 thermocouple thermometer (Everett, WA, USA) during exposure to LED illumination at doses as high as 576 J/cm2. The irradiance (W/cm2) of a 405 ± 5-nm LED unit was measured using a compact power and energy meter console (PM100D; Thorlabs GmbH, Dachau, Germany), with 20 ± 2 mW/cm2 at the surface of the cell suspension. The dosage applied to the sample was calculated as E = Pt, where E is the dose in J/cm2, P is irradiance in W/cm2, and t is time in s.
FIG 8.
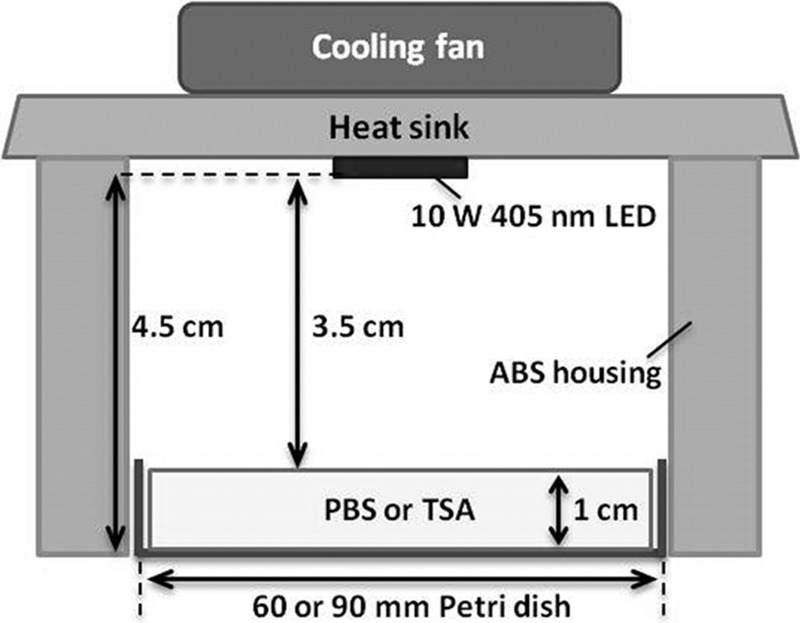
Schematic diagram of the 405 ± 5-nm LED illumination system.
Sensitivity of Salmonella to LED illumination.
To screen for sensitivity and resistance of Salmonella to LED illumination, a 10-μl bacterial suspension of each of the 18 Salmonella strains (ca. 106 CFU/ml) was streaked in individual lines ca. 3 cm in length using an inoculating loop onto the surface of TSA plates. The inoculated plates were placed in the LED system as previously described and exposed to 288 J/cm2 at a set temperature of 4°C in a temperature-controlled incubator (MIR-154; Panasonic Healthcare Co., Ltd., Osaka, Japan). Nonilluminated control plates were placed in a temperature-controlled incubator in the dark. After illumination, plates were incubated at 37°C for 24 h and then visualized using a G:Box EF2 imaging system (Syngene, Frederick, MD, USA).
LED illumination of the Salmonella suspension and enumeration.
Based on results from the above-described experiments, 10-ml suspensions of S. Enteritidis ATCC 13076 and S. Saintpaul at an initial population of ca. 106 CFU/ml were deposited in sterile petri dishes and placed in the LED illumination system, and then exposed to LED illumination at doses as high as 576 J/cm2 at a set temperature of 4°C. The same volume of cell suspension was placed in a sterile petri dish in an incubator without LED illumination (dark condition) to serve as a nonilluminated control. Aliquots of 0.5 ml in volume were taken at selected treatment times and analyzed for numbers of surviving Salmonella. Aliquots from the nonilluminated control and illuminated cell suspensions were serially diluted with 0.1% sterile peptone water (PW; Oxoid) and pour-plated in TSA. After incubating at 37°C for 24 to 48 h, the numbers of viable cells were enumerated using a colony counter (Rocker Scientific Co., Ltd., Taipei, Taiwan) and expressed as log CFU/ml. Bacterial survival curves were constructed by plotting log numbers of survivors against doses. The line of best fit for the survival curve was determined by linear regression with Microsoft Excel (Microsoft Corp., Redmond, WA, USA). The D values (J/cm2) for the first-order inactivation kinetics were calculated as the negative reciprocals of the slopes.
Determination of intracellular coproporphyrin content.
Cultures (100 ml) of S. Enteritidis ATCC 13076 and S. Saintpaul in stationary-growth phase in TSB were centrifuged at 6,000 × g for 10 min at 4°C, washed thrice with 20 ml of PBS, and dried at 45°C for 2 h. Intracellular coproporphyrin was extracted from the dried pellets with a 0.1 M NH4OH-acetone solution (1:9 [vol/vol]). The extracts were quantified using a C18 modified silica column, a reversed phase system, and a Waters 2695 HPLC system equipped with a multi-λ-fluorescent detector at 407 nm excitation and 612 nm emission. Elutions were carried out with a gradient separation consisting of water (solvent A) and acetonitrile (solvent B). A standard curve was prepared in a concentration range of 0 to 10 μg/ml of coproporphyrin I dihydrochloride (Sigma-Aldrich, St. Louis, MO, USA). The amount of coproporphyrin is expressed in ng coproporphyrin/mg dried cells.
Determination of cellular DNA oxidation.
The degree of cellular DNA oxidation caused by LED illumination was measured as described by Kim et al. (39). Briefly, suspensions (10 ml) of S. Enteritidis ATCC 13076 and S. Saintpaul (ca. 108 CFU/ml) were illuminated (576 J/cm2) at a set temperature of 4°C. Genomic DNA from LED-illuminated cells was extracted using a GeneJET genomic DNA purification kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. The concentration and purity of extracted genomic DNA dissolved in 100 μl of elution buffer were measured with a BioDrop DUO spectrophotometer (BioDrop, Cambridge, UK). Genomic DNA from freshly cultured cells and nonilluminated cells held at 9.5°C without LED illumination were extracted by the same method.
For the DNA oxidation assay, we used an OxiSelect oxidative DNA damage enzyme-linked immunosorbent assay (ELISA) kit (Cell Biolabs, San Diego, CA, USA). The DNA extract (1.3 to 3.1 μg) was converted to single-strand DNA at 95°C for 5 min, then immediately chilled in an ice bath and incubated with 6 units of nuclease PI (Wako, Osaka, Japan) in 20 mM sodium acetate buffer (Sigma-Aldrich) at 37°C for 2 h. Two units of Escherichia coli alkaline phosphatase (TaKaRa Bio, Inc., Shiga, Japan) were added to the mixture containing a 10% (vol/vol) 10× alkaline phosphatase buffer (TaKaRa Bio, Inc.) and incubated at 37°C for 1 h. The mixtures were centrifuged at 6,000 × g for 5 min at 4°C, and 8-hydroxydeoxyguanosine (8-OHdG) in the supernatant was quantified according to the manufacturer's instructions. The measurements were made at 450 nm with a Synergy HT multidetection microplate reader (BioTek Instruments, Inc., Winooski, VT, USA). The amount of 8-OHdG was calculated with a standard curve (0 to 10 ng/ml) and is reported as ng 8-OHdG per mg of DNA.
Determination of damage to cell membrane function.
To investigate damage to cell membrane functions by LED illumination, five fluorescent probes were used: SYTO9 and propidium iodide (PI) from the LIVE/DEAD BacLight kit (Molecular Probes, Thermo Fisher Scientific, Eugene, OR, USA), ethidium bromide (EtBr; Sigma-Aldrich), bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3); Molecular Probes], and 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy–d-glucose (2-NBDG; Molecular Probes). Ten-milliliter suspensions (ca. 106 CFU/ml) of S. Enteritidis ATCC 13076 and S. Saintpaul were exposed to 576 J/cm2 at a set temperature of 4°C; 0.5-ml or 1-ml aliquots were withdrawn at 288 and 578 J/cm2 of illumination, respectively. Aliquots (200 μl) were immediately stained with two mixed probes (SYTO9/PI or SYTO9/EtBr) and three single probes [SYTO9, DiBAC4(3), and 2-NBDG]. Before staining the cells, 2.0 mM 2,4-dinitrophenol (2,4-DNP; Sigma-Aldrich) was added to 2-NBDG. The working concentrations of SYTO9, PI, EtBr, DiBAC4(3), and 2-NBDG were 5, 30, 30, 10, and 5 μM, respectively. The mixtures were incubated in the dark for 10 min at 37°C for 2-NBDG and for 15 min at room temperature for SYTO9, PI, EtBr, and DiBAC4(3). The measurements were made using a BD Accuri C6 flow cytometer (Accuri Cytometers, Inc., Ann Arbor, MI, USA) equipped with a blue laser that excites at 488 nm and five detectors, including detectors of forward scatter (FSC) and side scatter (SSC), a green fluorescent filter (FL1; 533/30 nm), an orange fluorescent filter (FL2; 585/40 nm), and a red fluorescent filter (FL3; 670 nm). The detectors were used as follows: FL1 for SYTO9, FL3 for PI and EtBr, and FSC and SSC for DiBAC4(3) and 2-NBDG, respectively.
Transmission electron microscopy.
Ten petri dishes containing 10-ml suspensions (ca. 109 CFU/ml) of S. Enteritidis ATCC 13076 were individually placed in the LED system for illumination at a set temperature of 4°C at doses as high as 720 J/cm2. Ten sets of nonilluminated control cells were stored in the dark. A cell suspension pooled from 10 petri dishes was centrifuged at 6,000 × g for 10 min at 4°C to obtain a population of ca. 1010 CFU/ml. Cells in the resultant pellet were fixed with 2.5% (vol/vol) glutaraldehyde (Sigma-Aldrich) in PBS (pH 7.4) by incubating overnight at 4°C, and were then washed twice in 25 ml of PBS and stored at 4°C prior to TEM analysis. Cells were postfixed in 1% osmium tetroxide (OsO4; pH 7.4) for 2 h at room temperature, were washed twice with distilled water for 10 min, and were dehydrated through an ascending ethanol series (25% ethanol for 5 min, 50% ethanol for 10 min, 75% ethanol for 10 min, 95% ethanol for 10 min, and 100% ethanol for 10 min) and 100% acetone for 10 min at room temperature. The fixed cells were infiltrated with a 1:1 mixture of 100% acetone and Araldite resin (Pelco International, Redding, CA, USA) for 30 min and then infiltrated with a 1:6 mixture of 100% acetone and Araldite resin overnight at room temperature. Infiltrated samples were placed thrice in fresh Araldite resin for 1 h at 45°C, were embedded in fresh Araldite resin, and then cured at 60°C for 24 h. After curing, samples were sectioned and mounted on copper grids (200 mesh; Pelco International), and then stained with Reynolds' lead citrate (40). The stained samples were examined with a JEOL JEM 1010 transmission electron microscope (JEOL Asia, Ltd., Tokyo, Japan).
Total RNA isolation and real-time qRT-PCR.
After LED illumination (at 72 J/cm2) of S. Enteritidis ATCC 13076 and S. Saintpaul at a set temperature of 4°C, the total RNA in illuminated cells was isolated with an RNeasy minikit (Qiagen, Inc., Hilden, Germany) according to the manufacturer's instructions. The total RNAs in freshly cultured cells and nonilluminated cells stored at 9.5°C without LED illumination were also extracted by the same method. The purified RNA was dissolved in 50 μl of RNase-free water. The purity and the concentration of extracted RNA were determined using a BioDrop Touch dual spectrophotometer. The integrity of total RNA was evaluated by 1% (wt/vol) agarose gel electrophoresis.
One microgram of total RNA was converted to cDNA using a QuantiTect reverse transcription kit (Qiagen, Inc.) according to the manufacturer's instructions. Real-time quantitative reverse transcription-PCR (qRT-PCR) was carried out with a StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) to determine the relative expression levels of oxyR, recA, rpoS, sodA, and soxR in nonilluminated and illuminated cells. All primers used in this study were designed using Primer Expression Software 3.0 (Applied Biosystems) based on the nucleotide sequence derived from the NCBI database (accession number NC_011294.1). 16S rRNA was used as a reference gene, and the primer sequence was obtained from Yang et al. (5). Detailed information concerning these target genes and primer sequences is presented in Table 2. The reaction mixtures (20 μl) contained 10 μl of Fast SYBR green master mix (Applied Biosystems), 1 μl of 0.5 μM forward primer, 1 μl of 0.5 μM reverse primer, 1 μl of template cDNA, and 7 μl of PCR water. Thermal cycling conditions consisted of 1 cycle at 95°C for 20 s, followed by 40 cycles at 95°C for 3 s and 60°C for 30 s. The absence of DNA contamination as a negative control was confirmed by reactions without reverse transcriptase. The changes in relative gene expression were calculated with the 2–ΔΔCT method (5).
TABLE 2.
Gene functions and primer sequences
| Gene | Functiona | Primer sequence (5′ to 3′)b | Product size |
|---|---|---|---|
| oxyR | Regulates hydrogen peroxide detoxification | F:ACCCTTAGCGGGCAAATACG | 62 bp |
| R:CGTACGCTCCAGCAGCATAA | |||
| sodA | Encodes manganese superoxide dismutase | F:CCCTGCCGTACGCTTATGA | 89 bp |
| R:TGACATAGGTTTGATGGTGTTTGG | |||
| soxR | Regulates resistance to superoxide-generating agents | F:GGCGACGCGTTTGGTATC | 73 bp |
| R:CACTGCGAGGAGAGCTGCTT | |||
| recA | Induces SOS response and repairs DNA damage | F:TCGCCGTTGTAGCTGTACCA | 65 bp |
| R:GGTTGACCTGGGCGTGAA | |||
| rpoS | Regultes general stress responses (e.g., to carbon starvation, acid stress, oxidative stress, etc.) | F:CCACCAGGTTGCGTATGTTG | 63 bp |
| R:CCGTGCAGTCGAGAAGTTTG |
Functions as described by Farr and Kogoma (13).
F, forward; R, reverse.
Statistical analysis.
All mean values for each of the data points were obtained by performing independent triplicates with duplicate analyses (n = 6). All data are expressed as means ± the standard deviations, and the significant differences between mean values were calculated at the 95% confidence interval with analyses of variance (ANOVAs) and least significant difference (LSD) tests using SPSS statistical software (version 20; IBM Corp., Armonk, NY, USA).
ACKNOWLEDGMENTS
This study was supported in part by funding from the Ministry of Education's Academic Research Fund (AcRF) Tier 1 (R-143-000-624-112). We thank L. R. Beuchat at the University of Georgia for his thoughtful review of the manuscript and K. Seo at Konkuk University for kindly providing S. Enteritidis strains.
The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Lathrop S. 2015. Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2014. MMWR Morb Mortal Wkly Rep 64:495–499. [PMC free article] [PubMed] [Google Scholar]
- 3.Kondakci T, Yuk HG. Dec 2012/Jan 2013 Overview of foodborne outbreaks in the last decade in Singapore: alarming increase in nontyphoidal salmonellosis, p 42–45. In Food and beverage Asia. http://www.foodbeverageasia.com/ebook/FBA_DecJan2013/files/assets/seo/page44.html.
- 4.Ivancic T, Jamnik P, Stopar D. 2013. Cold shock CspA and CspB protein production during periodic temperature cycling in Escherichia coli. BMC Res Notes 6:248. doi: 10.1186/1756-0500-6-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Khoo WJ, Zheng Q, Chung HJ, Yuk HG. 2014. Growth temperature alters Salmonella Enteritidis heat/acid resistance, membrane lipid composition and stress/virulence related gene expression. Int J Food Microbiol 172:102–109. doi: 10.1016/j.ijfoodmicro.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Luksienė Z, Zukauskas A. 2009. Prospects of photosensitization in control of pathogenic and harmful micro-organisms. J Appl Microbiol 107:1415–1424. doi: 10.1111/j.1365-2672.2009.04341.x. [DOI] [PubMed] [Google Scholar]
- 7.Luksiene Z, Paskeviciute E. 2011. Novel approach to the microbial decontamination of strawberries: chlorophyllin-based photosensitization. J Appl Microbiol 110:1274–1283. doi: 10.1111/j.1365-2672.2011.04986.x. [DOI] [PubMed] [Google Scholar]
- 8.Maclean M, MacGregor SJ, Anderson JG, Woolsey G. 2009. Inactivation of bacterial pathogens following exposure to light from a 405-nanometer light-emitting diode array. Appl Environ Microbiol 75:1932–1937. doi: 10.1128/AEM.01892-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghate VS, Ng KS, Zhou W, Yang H, Khoo GH, Yoon WB, Yuk HG. 2013. Antibacterial effect of light emitting diodes of visible wavelengths on selected foodborne pathogens at different illumination temperatures. Int J Food Microbiol 166:399–406. doi: 10.1016/j.ijfoodmicro.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Kim MJ, Mikš-Krajnik M, Kumar A, Ghate V, Yuk HG. 2015. Antibacterial effect and mechanism of high-intensity 405 ± 5 nm light emitting diode on Bacillus cereus, Listeria monocytogenes, and Staphylococcus aureus under refrigerated condition. J Photochem Photobiol B 153:33–39. doi: 10.1016/j.jphotobiol.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Kim MJ, Mikš-Krajnik M, Kumar A, Yuk HG. 2016. Inactivation by 405 ± 5 nm light emitting diode on Escherichia coli O157:H7, Salmonella Typhimurium, and Shigella sonnei under refrigerated condition might be due to the loss of membrane integrity. Food Control 59:99–107. doi: 10.1016/j.foodcont.2015.05.012. [DOI] [Google Scholar]
- 12.Kumar A, Ghate V, Kim MJ, Zhou W, Khoo GH, Yuk HG. 2015. Kinetics of bacterial inactivation by 405 nm and 520 nm light emitting diodes and the role of endogenous coproporphyrin on bacterial susceptibility. J Photochem Photobiol B 149:37–44. doi: 10.1016/j.jphotobiol.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev 55:561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamblin MR, Hasan T. 2004. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci 3:436–450. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen R, van der Straaten T, van Diepen A, van Dissel JT. 2003. Responses to reactive oxygen intermediates and virulence of Salmonella typhimurium. Microbes Infect 5:527–534. doi: 10.1016/S1286-4579(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 16.Michán C, Manchado M, Dorado G, Pueyo C. 1999. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J Bacteriol 181:2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh TJ, Kim IG. 1999. Identification of genetic factors altering the SOS induction of DNA damage-inducible yebG gene in Escherichia coli. FEMS Microbiol Lett 177:271–277. doi: 10.1111/j.1574-6968.1999.tb13743.x. [DOI] [PubMed] [Google Scholar]
- 19.Berney M, Weilenmann HU, Egli T. 2006. Flow-cytometric study of vital cellular functions in Escherichia coli during solar disinfection (SODIS). Microbiology 152:1719–1729. doi: 10.1099/mic.0.28617-0. [DOI] [PubMed] [Google Scholar]
- 20.Endarko E, Maclean M, Timoshkin IV, MacGregor SJ, Anderson JG. 2012. High-intensity 405 nm light inactivation of Listeria monocytogenes. Photochem Photobiol 88:1280–1286. doi: 10.1111/j.1751-1097.2012.01173.x. [DOI] [PubMed] [Google Scholar]
- 21.Murdoch LE, Maclean M, Endarko E, MacGregor SJ, Anderson JG. 2012. Bactericidal effects of 405 nm light exposure demonstrated by inactivation of Escherichia, Salmonella, Shigella, Listeria, and Mycobacterium species in liquid suspensions and on exposed surfaces. ScientificWorldJournal 2012:137805. doi: 10.1100/2012/137805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gayán E, Serrano MJ, Raso J, Álvarez I, Condón S. 2012. Inactivation of Salmonella enterica by UV-C light alone and in combination with mild temperatures. Appl Environ Microbiol 78:8353–8361. doi: 10.1128/AEM.02010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamamoto A, Mori M, Takahashi A, Nakano M, Wakikawa N, Akutagawa M, Kinouchi Y. 2007. New water disinfection system using UVA light-emitting diodes. J Appl Microbiol 103:2291–2298. doi: 10.1111/j.1365-2672.2007.03464.x. [DOI] [PubMed] [Google Scholar]
- 24.Salmon-Divon M, Nitzan Y, Malik Z. 2004. Mechanistic aspects of Escherichia coli photodynamic inactivation by cationic tetra-meso (N-methylpyridyl) porphine. Photochem Photobiol Sci 3:423–429. doi: 10.1039/b315627n. [DOI] [PubMed] [Google Scholar]
- 25.Amaral L, Martins A, Spengler G, Molnar J. 2014. Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol 4:168. doi: 10.3389/fphar.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christena LR, Mangalagowri V, Pradheeba P, Ahmed KBA, Shalini BIS, Vidyalakshmi M, Anbazhagan V. 2015. Copper nanoparticles as an efflux pump inhibitor to tackle drug resistant bacteria. RSC Adv 5:12899–12909. doi: 10.1039/C4RA15382K. [DOI] [Google Scholar]
- 27.Thöny-Meyer L. 2009. Heme transport and incorporation into proteins, p 149–159. In Warren MJ, Smith AG (ed), Tetrapyrroles: birth, life and death. Springer Science and Business Media, New York, NY. [Google Scholar]
- 28.Nitzan Y, Ashkenazi H. 2001. Photoinactivation of Acinetobacter baumannii and Escherichia coli B by a cationic hydrophilic porphyrin at various light wavelengths. Curr Microbiol 42:408–414. doi: 10.1007/s002840010238. [DOI] [PubMed] [Google Scholar]
- 29.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reizer J, Saier MH, Deutscher J, Grenier F, Thompson J, Hengstenberg W, Dills SS. 1988. The phosphoenolpyruvate: sugar phosphotransferase system in Gram-positive bacteria: properties, mechanism, and regulation. Crit Rev Microbiol 15:297–338. doi: 10.3109/10408418809104461. [DOI] [PubMed] [Google Scholar]
- 31.Caminos DA, Spesia MB, Pons P, Durantini EN. 2008. Mechanisms of Escherichia coli photodynamic inactivation by an amphiphilic tricationic porphyrin and 5,10,15,20-tetra(4-N,N,N-trimethylammoniumphenyl) porphyrin. Photochem Photobiol Sci 7:1071–1078. doi: 10.1039/b804965c. [DOI] [PubMed] [Google Scholar]
- 32.Alves E, Faustino MA, Neves MG, Cunha A, Tome J, Almeida A. 2014. An insight on bacterial cellular targets of photodynamic inactivation. Future Med Chem 6:141–164. doi: 10.4155/fmc.13.211. [DOI] [PubMed] [Google Scholar]
- 33.Spesia MB, Caminos DA, Pons P, Durantini EN. 2009. Mechanistic insight of the photodynamic inactivation of Escherichia coli by a tetracationic zinc(II) phthalocyanine derivative. Photodiagnosis Photodyn Ther 6:52–61. doi: 10.1016/j.pdpdt.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Smirnova GV, Zakirova ON, Oktyabrskii ON. 2001. The role of antioxidant systems in the cold stress response of Escherichia coli. Microbiology 70:45–50. doi: 10.1023/A:1004840720600.11338835 [DOI] [Google Scholar]
- 35.Limthammahisorn S, Brady YJ, Arias CR. 2008. Gene expression of cold shock and other stress-related genes in Vibrio vulnificus grown in pure culture under shellstock temperature control conditions. J Food Prot 71:157–164. doi: 10.4315/0362-028X-71.1.157. [DOI] [PubMed] [Google Scholar]
- 36.Kong IS, Bates TC, Hülsmann A, Hassan H, Smith BE, Oliver JD. 2004. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol 50:133–142. doi: 10.1016/j.femsec.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST. 2009. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc Natl Acad Sci U S A 106:611–616. doi: 10.1073/pnas.0803665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Child M, Strike P, Pickup R, Edwards C. 2002. Salmonella Typhimurium displays cyclical patterns of sensitivity to UV-C killing during prolonged incubation in the stationary phase of growth. FEMS Microbiol Lett 213:81–85. doi: 10.1111/j.1574-6968.2002.tb11289.x. [DOI] [PubMed] [Google Scholar]
- 39.Kim MJ, Bang WS, Yuk HG. 2017. 405 ± 5 nm light emitting diode illumination causes photodynamic inactivation of Salmonella spp. on fresh-cut papaya without deterioration. Food Microbiol 62:124–132. doi: 10.1016/j.fm.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]