Figure 4.
The Levels of CtIP During Reprogramming Affect iPSC Maintenance
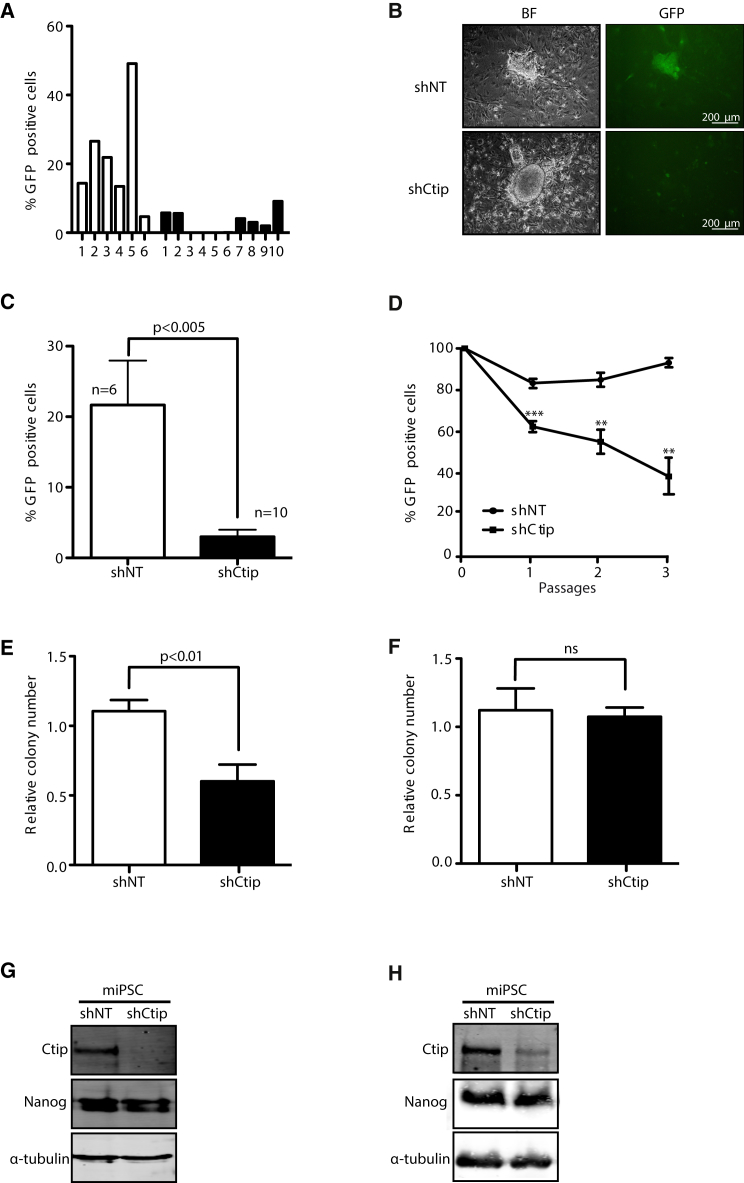
(A) Percentage of GFP-positive cells in iPSC colonies isolated after MEFs reprogramming in the presence (white bars) or absence (black bars) of CtIP.
(B) Representative bright field (BF) and GFP images of miPSCs obtained by microscopy.
(C) Average percentage of GFP cells in several clones of shNT and shCtIP iPSCs. Error bars indicate ±SEM of different clones (n = 6 and n = 10). Statistical analysis was performed using a t test.
(D) Percentage of iPSCs harboring shCtIP-GFP or shNT-GFP during subsequence passages of the cells. The average and SD of five independent experiments is plotted. t test analysis for each passage is shown. ∗∗p < 0.01, ∗∗∗p < 0.005.
(E) The same amount of miPSCs reprogrammed either in the presence (white bars) or absence (black bars) of CtIP were seeded at low density and the number of each colony was measured. The relative number of colonies formed from three independent experiments is plotted. The t test was performed to compare both conditions.
(F) Same as (E) but with cells reprogrammed in the presence of CtIP, and then transduced with an shRNA against CtIP (black bars) or control shNT (white bars). Other details as in (E). ns, not significant.
(G) miPSCs reprogrammed in the presence of shCtIP or control shNT were immunoblotted to analyze the indicated proteins. A representative western blot is shown of three independent experiments.
(H) Same as in (G) but with cells transduced with an shRNA against CtIP after reprogramming.
See also Figures S4 and S5.