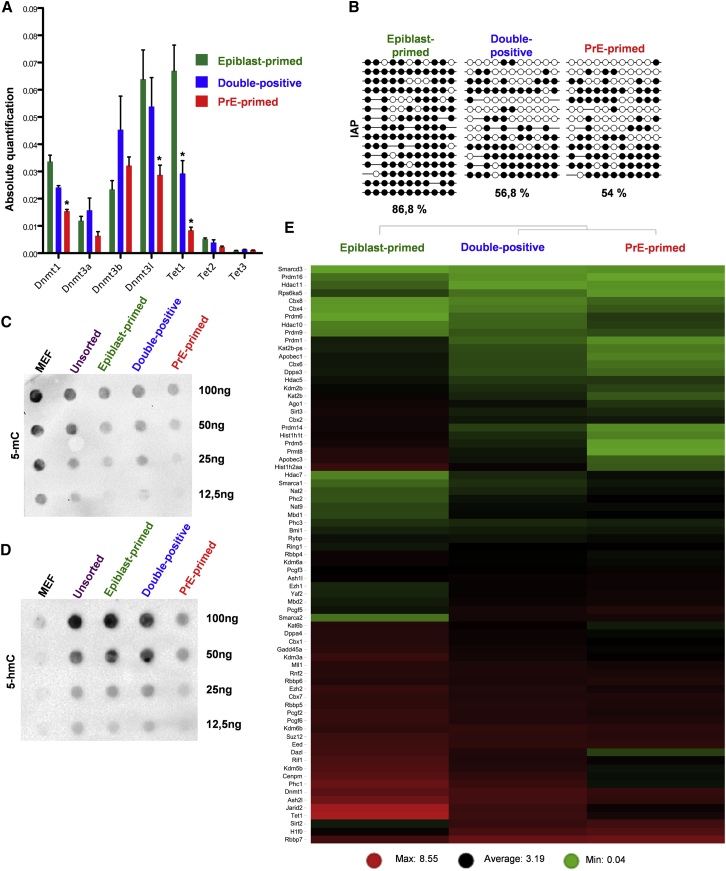
Figure 4.
Different Epigenetic State of PDGFRα+ Subpopulations
(A) qRT-PCR analysis for DNA methylation/hydroxymethylation genes upon sorting of the respective subpopulations. Data are presented as means ± SEM of each transcript from three independent experiments (normalized to β-Actin), ∗p < 0.05, t test.
(B) Bisulfite sequencing of IAP sequences. Open circles, unmethylated; closed circles, methylated.
(C) Representative dot blot for global 5-mC, from three independent experiments. Mouse embryonic fibroblasts (MEF) were used as control as they contain high levels of 5-mC and low levels of 5-hmC.
(D) Representative dot blot for global 5-hmC from three independent experiments. MEFs were used as control as they contain low levels of 5-hmC.
(E) Heatmap of epigenetic regulators on sorted subpopulations based on RNA-seq data. Transcript levels are based on FPKM (fragments per kilobase of exon per million fragments mapped). Red and green represent high and low gene expression, respectively.