Abstract
Precise regulation of mRNA processing, translation, localization, and stability relies on specific interactions with RNA-binding proteins whose biological function and target preference are dictated by their preferred RNA motifs. The RBPMS family of RNA-binding proteins is defined by a conserved RNA recognition motif (RRM) domain found in metazoan RBPMS/Hermes and RBPMS2, Drosophila couch potato, and MEC-8 from Caenorhabditis elegans. In order to determine the parameters of RNA sequence recognition by the RBPMS family, we have first used the N-terminal domain from MEC-8 in binding assays and have demonstrated a preference for two GCAC motifs optimally separated by >6 nucleotides (nt). We have also determined the crystal structure of the dimeric N-terminal RRM domain from MEC-8 in the unbound form, and in complex with an oligonucleotide harboring two copies of the optimal GCAC motif. The atomic details reveal the molecular network that provides specificity to all four bases in the motif, including multiple hydrogen bonds to the initial guanine. Further studies with human RBPMS, as well as Drosophila couch potato, confirm a general preference for this double GCAC motif by other members of the protein family and the presence of this motif in known targets.
Keywords: ITC, NMR, X-ray crystallography, protein–RNA, structure
INTRODUCTION
The regulation of mRNA processing involves protein factors that recognize specific RNA elements in order to modulate various properties of the mature mRNA. RNA-binding proteins mediate transport of the final mRNA to the cytoplasm and can alter its stability or define a precise sub-cellular localization. Initial processing steps of the pre-mRNA are also targets of specific protein factors. During splicing, the definition of intron boundaries involves specific recognition of intronic RNA sequences by constitutive splicing factors. An additional set of protein factors can be expressed during development or in response to an external stimulus to alter the inclusion or exclusion of exons to change mRNA stability and function, as well as the function of the encoded protein isoforms. In order to understand the mechanism and target selection by these RNA-binding proteins, both the protein factors and RNA motif elements need to be characterized at the molecular level.
One set of RNA-binding proteins, the RBPMS family, is named after the vertebrate RNA-binding protein gene with multiple splicing (RBPMS) (Shimamoto et al. 1996). The first member cloned was the couch potato gene from Drosophila (Bellen et al. 1992a) followed by the mec-8 gene from C. elegans (Lundquist et al. 1996). Vertebrate RBPMS and the paralog RBPMS2 are expressed in several tissues including heart, muscle, kidney, liver, lung, eye, adipose, and ovary (Wilmore et al. 2005; Kwong et al. 2010; Farazi et al. 2014). RBPMS affects synapse density and axon formation in the retina ganglion cells (Piri et al. 2006; Hornberg et al. 2013; Rodriguez et al. 2014) and colocalizes with NonO, PSF, and G3BP1 in mouse neuron cells (Furukawa et al. 2015). Expression of RBPMS2 in the intestine may play a role in motility disorders (Notarnicola et al. 2012) and gastrointestinal tumors (Hapkova et al. 2013). The connection to cancer and cell cycle control is also suggested by an interaction with cFOS to regulate AP-1 target genes (Fu et al. 2015). A role in heart development was observed for chicken RBPMS and the Xenopus orthologs (Gerber et al. 1999, 2002) and Xenopus RBPMS/Hermes is further implicated in early oocyte maturation (Chang et al. 2004; Zearfoss et al. 2004; Song et al. 2007; Aguero et al. 2016).
The founding member of the RBPMS family is couch potato from Drosophila, originally identified in a genetic screen for components involved in neuron differentiation and in particular the sensory organs of the peripheral nervous system (Bellen et al. 1992b). Similar to human RBPMS, the couch potato gene exhibits alternative splicing and is relatively large (>100 kb). Flies with a mutant couch potato allele are less active (Bellen et al. 1992a,b) and display other neurological defects (Glasscock and Tanouye 2005). In addition, natural variation in the gene may underlie seasonal and geographical adaptation to environmental stress in several Drosophila species (Schmidt et al. 2008; Salminen et al. 2015).
Finally, the RBPMS family member in C. elegans, mec-8 (mechanosensory abnormality gene 8), was first described following the characterization of touch-insensitive mutants (Chalfie and Sulston 1981). Subsequently, MEC-8 was found to regulate the alternative splicing of the perlecan homolog unc-52 in the hyperdermis (Lundquist and Herman 1994; Spike et al. 2002) and an isoform of the stomatin homolog mec-2 required for function of the six mechanosensory neurons (Calixto et al. 2010).
At the molecular level, all members of the RBPMS family share a conserved RNA recognition motif (RRM) domain (Fig. 1A). Previous work has established that the RRM domains from RBPMS2, RBPMS, and Xenopus RBPMS/Hermes form homodimers in solution (Sagnol et al. 2014; Aguero et al. 2016; Teplova et al. 2016). Disruption of the domain interface in chicken RBPMS2 abolished normal protein function related to intestinal development, and in vitro the mutant protein showed reduced binding to a segment of NOGGIN mRNA (Sagnol et al. 2014). Dimerization mutants of human RBPMS failed to localize to arsenite-triggered stress granules, and similar to RBPMS2, reduced RNA binding was observed by the mutant (Teplova et al. 2016). Although the residues involved in dimerization are further conserved in MEC-8 and couch potato, the oligomeric state of these proteins had not been investigated.
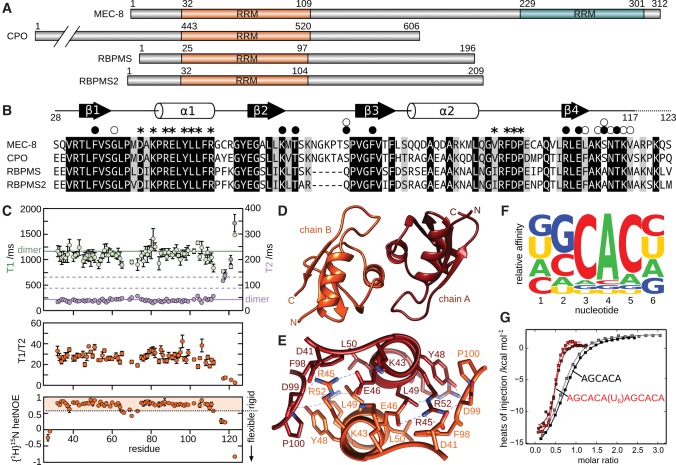
FIGURE 1.
Characterization of MEC-8 RRM1 dimer and ligand sequence. (A) RNA recognition motif (RRM) domains within C. elegans MEC-8 (UniProt G5ECJ4), Drosophila melanogaster couch potato (CPO; UniProt Q01617), human RBPMS (UniProt Q93062), and human RBPMS (UniProt Q6ZRY4). Residue numbers are indicated. Note that all four proteins exist as multiple splice isoforms, with only the N-terminal RRM domain common to all MEC-8, couch potato, and RBPMS variants. (B) Sequence alignment of the conserved RRM domain of the RBPMS family. Regions of β-strands (black arrows) and α-helices (white cylinders) are indicated above the MEC-8 sequence, based on the crystal structure of the unbound MEC-8 RRM1 domain. Residues involved in dimerization are indicated with asterisks. Residue sidechains involved in ligand binding are indicated with a black circle, and interactions with backbone atoms have a white circle. Identical residues in all four sequences are highlighted in black and those that are homologous by sidechain properties are shaded in gray. (C) NMR spectroscopy characterization based on residue-specific 15N relaxation measurements of T1 (light green), T2 (purple), the combined ratio of T1/T2 and {1H}15N heteronuclear NOE. Predicted values of monomeric and dimeric RRM1 are indicated by dashed and solid lines, respectively (Daragan and Mayo 1997). Values of {1H}15N heteronuclear NOE below 0.6 correspond to backbone amides with significant conformational disorder. (D,E) Structure overview and close-up of the dimerization interface of the unbound MEC-8 RRM1 dimer (PDB 5BJR). (F) Relative affinity of ligands with individual nucleotide substitutions binding to MEC-8 RRM1 by using isothermal titration calorimetry (ITC). The heights of the A, C, G, and U in each position are relative to the determined values of the association constant, KA. Representative raw data and derived KA values can be found in Supplemental Figure S2. (G) Representative data from ITC measurements of MEC-8 RRM1 with the RNA ligands AGCACA and AGCACAUUUUUUAGCACA.
In terms of the RNA sequences recognized by RBPMS, a PAR-CLIP approach identified RNA targets with multiple CAC motifs separated by varying lengths (Farazi et al. 2014). Additional electrophoresis mobility shift assays demonstrated in vitro interaction of RBPMS and RBPMS2 with AC-rich RNA ligands, as well as oligonucleotides based on wild-type and mutated segments of mRNA from UBE2V1, SRM, ETF1, and NDUA6. A crystal structure of the RRM domain of human RBPMS in complex with a short RNA confirmed recognition of the trinucleotide CAC (Teplova et al. 2016). In contrast to these observations, a complementary approach involving RNA-compete identified a longer 4-nucleotide (nt) GCAC motif recognized by other members of the RBPMS family, namely couch potato from Drosophila and MEC-8 from C. elegans (Ray et al. 2013).
In this study, we use crystal structures and binding data from isothermal titration calorimetry to show that the N-terminal RRM domain from MEC-8 is a dimer and interacts with highest affinity to a double GCAC motif. Additional experiments on the RRM domains from couch potato and RBPMS demonstrate shared binding preferences by this family of RNA-binding proteins.
RESULTS AND DISCUSSION
Based on a sequence alignment of MEC-8, Drosophila couch potato, human RBPMS and human RBPMS2 (Fig. 1B), we have cloned the first RRM domain (RRM1) from MEC-8 as a construct from residues 28–123 (hereafter MEC-8 RRM1). Characterization of the purified protein by NMR spectroscopy reveals that MEC-8 RRM1 forms a stable and soluble dimer in solution, similar to the situation with human RBPMS and RBPMS2 (Sagnol et al. 2014; Teplova et al. 2016). Specifically, NMR spectroscopy allows for the estimate of molecular size from measurement of the relaxation properties of the amide nitrogens in the protein backbone. The T1 and T2 15N relaxation values (Fig. 1C) are consistent with the values predicted for a dimer (solid lines) as opposed to values for a monomer (dotted lines) (Daragan and Mayo 1997). The measurements also allow for an approximate calculation of molecular size by using the ratio of T1 over T2 to give an average value of 26 ± 7 kDa, again close to the calculated dimer mass of 22 kDa. As a final characterization by NMR spectroscopy, we looked at conformational disorder of the protein backbone by measuring 15N heteronuclear NOE, which reveals that residues from Val117 to the C terminus are highly flexible (values <0.6). We therefore made a second construct in which the last six residues were removed (MEC-8 RRM128-117) without otherwise perturbing the structure or dimerization. This second construct was successful in producing a crystal that diffracted to 2.6 Å resolution (complete statistics in Materials and Methods). The atomic details of the MEC-8 RRM128-117 dimer reveal an overall interface involving residues Asp41, Lys43, Arg45, Glu46, Tyr48, Leu49, Leu50, and Arg52 of helix α1, and residues Val96, Phe98, Asp99, and Pro100 from the loop between helix α2 and strand β4 (Fig. 1E). This dimer interface is consistent with that observed for both human RBPMS (Teplova et al. 2016) and human RBPMS2 (Sagnol et al. 2014) and confirms that MEC-8 forms a similar dimer arrangement to other members of the RBPMS family. Furthermore, we did not observe any evidence of a monomer in the concentrations used for NMR spectroscopy or ITC, and in contrast to RBPMS and RPBMS2, attempts to disrupt dimerization via comparable interface mutations (Arg45Glu or Leu49Glu) instead resulted in a loss of the protein fold (Supplemental Fig. S1). Dimer formation for MEC-8 may therefore be required, at least in vitro, for stabilizing the RRM1 domains.
In order to obtain atomic information on RNA binding by MEC-8 RRM1, an initial aim was to define the RNA sequence elements necessary for a high affinity ligand. The first goal was to define the optimal RNA binding sequence for one monomer of the MEC-8 RRM1 dimer. The MEC-8 protein was previously found to recognize a GCAC motif based on the RNA compete method (Ray et al. 2013) and a shorter CAC motif was found to be enriched in targets of other RBPMS family members by using the PAR-CLIP approach (Farazi et al. 2014). To better define the affinity of various sequences to MEC-8 RRM1, we used isothermal titration calorimetry (ITC) and a series of 6-nt ligands derived from a central GCAC motif. Individual measurements used ligands with all four bases at the 5′ and 3′ positions (Amrane et al. 2014). In addition, each of the GCAC bases was sequentially mutated to the other three bases in order to determine the importance of each nucleotide in the motif. The resulting ITC measurements therefore illustrate the relative effect on affinity for each base in the six positions (Fig. 1F; Supplemental Fig. S2). From these results, MEC-8 RRM1 does not appear to have selectivity for the first or last position in the hexamer sequences. In contrast, the CAC motif is clearly required for high affinity binding, preceded by a moderate preference for guanine in the second position of the tested ligands.
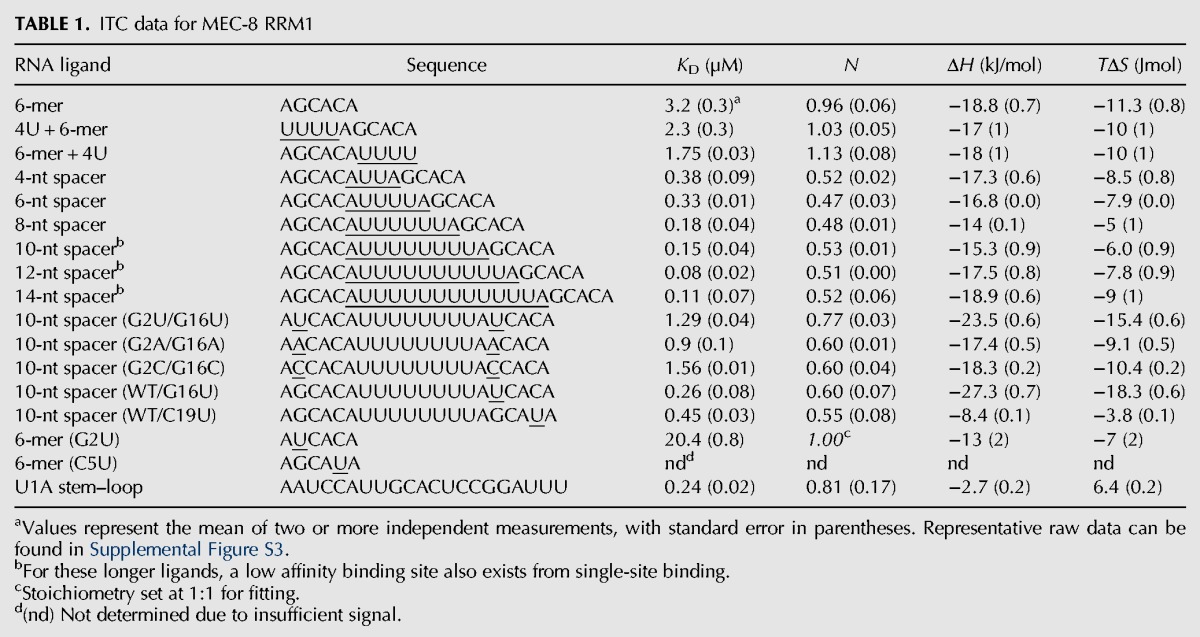
The dimeric nature of MEC-8 RRM1 adds another important aspect to RNA recognition, such that the highest affinity ligands are expected to contain two copies of this GCAC motif. We therefore used ITC to test the dimeric MEC-8 RRM1 with a series of longer RNA ligands in which two GCAC motifs are separated by 4–14 nt (Table 1; Supplemental Fig. S3). Simple extension of the AGCACA 6-mer by four uracils to the 5′ or 3′ end resulted in only minor changes in affinity (2.3 ± 0.3 µM and 1.75 ± 0.03 µM, respectively, as compared to 3.2 ± 0.3 µM), whereas all of the double motif ligands from the 4-nt spacer to the 14-nt spacer bound with higher affinity than the KD for the single GCAC motif. As expected, the stoichiometry of the complexes shifted from a 1:1 binding to a 1:2 ratio, with one RNA ligand recognized by a dimer of MEC-8 RRM1 (Fig. 1G; a 0.5:1 ratio of RNA ligand per MEC-8 monomer, versus 1:1 for the shorter AGCACA). The highest affinity ligands contain a linker of eight or more bases (KD values of 80–150 nM) and we therefore prepared crystallization screens with a variety of these oligonucleotides.
TABLE 1.
ITC data for MEC-8 RRM1
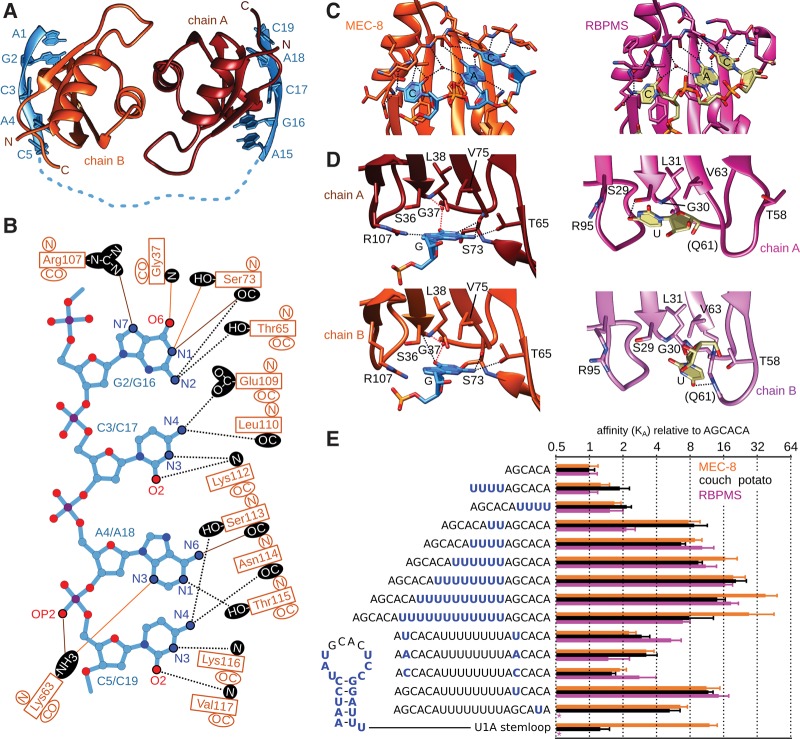
Successful crystallization and structure determination at 1.53 Å resolution was obtained for a complex containing the shorter MEC-8 RRM1 construct (residues 28–117) and the DNA version of the 10-nt spacer ligand (AGCACATTTTTTTTAGCACA). The overall structure and domain orientation of the MEC-8 RRM128-117 dimer (Fig. 2A) is similar to the arrangement of the unbound structure (Fig. 1D), with an rmsd of 0.6 Å. Density was observed for nucleotides that correspond to each of the two AGCAC sequences within the ligand. In contrast, nine linker nucleotides are not visible, nor is the 3′ terminal adenine. Due to crystal packing that places the adjacent complex against the first adenine, it was possible to unambiguously assign the two oligonucleotide stretches within the complex.
FIGURE 2.
Molecular basis of GCAC recognition by MEC-8 RRM1. (A) Side view of the MEC-8 RRM1(28–117) dimer bound to AGCACATTTTTTTTAGCACA (PDB 5TKZ). (B) Summary of hydrogen bonds between each MEC-8 RRM1 domain with the nucleotide ligand. Shared hydrogen bonds are indicated with black dotted lines. Hydrogen bonds specific to the first or second GCAC are indicated with orange or brown lines, respectively. (C) Common molecular recognition of the CAC trinucleotide by MEC-8 RRM1(28–117) (orange; chain B) and RBPMS (magenta; chain A of PDB 5DET) (Teplova et al. 2016). Hydrogen bonds are shown as dashed lines. (D) Close-up of the molecular details of guanine binding by MEC-8 and the uracil binding by RBPMS, colored as in C. (E) RNA binding affinity measured by isothermal titration calorimetry (ITC) for C. elegans MEC-8 RRM1, D. melanogaster couch potato(447-543), and human RBPMS(11–111) normalized for each protein by the KD of the 6-mer, and with error as the root mean square deviation.
In terms of atomic details, Ade1 and Ade15 stack against the following guanine but do not otherwise directly contact the protein surface. In contrast, specific binding of Gua2 or Gua16 is due to four hydrogen bonds each to MEC-8. A common set of hydrogen bonds connects the guanine N2 atom to both the backbone carbonyl of Ser73 and the sidechain hydroxyl of Thr65 (Fig. 2B). Due to a slight change in conformation between the bound guanines, the N1 atoms in Gua2 and Gua16 interact with either the backbone carbonyl or sidechain hydroxyl of Ser73, respectively. Another difference is that Gua2 has a hydrogen bond between N7 and the sidechain of Arg107, whereas in Gua16 this fourth hydrogen bond is between O2 and the backbone amide of Gly37. For the remaining CAC trinucleotide, the contacts are mainly similar on each MEC-8 RRM1 monomer. Cyt3/Cyt17 stack on Phe34 and are further stabilized by hydrogen bonds to the sidechain of Glu109 and backbone atoms of Leu110 and Lys112. Ade4/Ade18 stack on Phe77 with a hydrogen bond to the sidechain of Thr115. In addition, a second hydrogen bond is formed to either the backbone carbonyl of Ser113 for Ade4 or the sidechain of Lys63 for Ade18. Finally, Cyt5/Cyt19 stack onto the preceding adenine and are stabilized by the same four hydrogen bonds to the sidechain hydroxyl of Ser113, the backbone carbonyl of Asn114 and the backbone amides of Lys116 and Val117.
A comparison with the previously determined structure of RBPMS bound to a UCAC sequence (Teplova et al. 2016) reveals a high similarity in the recognition mode of the common CAC trinucleotide (Fig. 2C). In contrast, there is a significant difference in the position and degree of hydrogen bonding toward the initial guanine in the MEC-8 complex versus the corresponding uracil in the structure of bound RBPMS (Fig. 2D). In RBPMS, the uracil in each model is stabilized by only one direct hydrogen bond (<3.4 Å) as compared to the four hydrogen bonds between the guanine and residues within MEC-8. This raises the question as to the general importance of the guanine in the RNA motif recognized by MEC-8 RRM1 and for the rest of the RBPMS family. To address this aspect, we first determined the effect of mutating both guanine bases in the 10-nt-spacer RNA ligand. Mutation of both guanine bases to uracil (UCAC), adenine (ACAC), or cytosine (CCAC) reduced the affinity by a factor of six to ten (KD values of 1.29 ± 0.04 µM, 0.9 ± 1.56 µM and 1.56 ± 0.01 µM, respectively, as compared to a KD of 0.15 ± 0.04 µM for the ligand with guanine; Table 1). In contrast, maintaining at least one GCAC motif ensures relatively high affinity even when the second motif was mutated to the moderately or severely reduced-binding UCAC or GCAU sequences, respectively (Table 1).
To determine whether the RNA binding preferences exhibited by MEC-8 RRM1 were shared with other members of the RBPMS family, we repeated the ITC experiments with the RRM domains from D. melanogaster couch potato (residues 447–543) and human RBPMS (residues 11–111). Starting with the tandem GCAC ligands, both couch potato and RBPMS RRM domains displayed highest affinity when the two motifs were separated by 10–12 nt (Table 2; Supplemental Figs. S4, S5). The common mode of binding between these three members of the RBPMS family is clear when the individual KD values are normalized for each protein based on the affinity to the 6-mer AGCACA ligand (Fig. 2E). This binding similarity also extends to thermodynamic trends for the spacer-length series of RNA ligands. For all three proteins, the enthalpy of binding (ΔH) becomes less exothermic from the 4-nt to the 8-nt spacer ligand (Tables 1, 2), at which point it reverses and increasing linker length binds with more negative ΔH. This pattern is countered by the entropic term (TΔS) which is most unfavorable for the shortest and longest ligands and highest for the 8-nt linker.
TABLE 2.
ITC data for couch potato RRM and RBPMS RRM
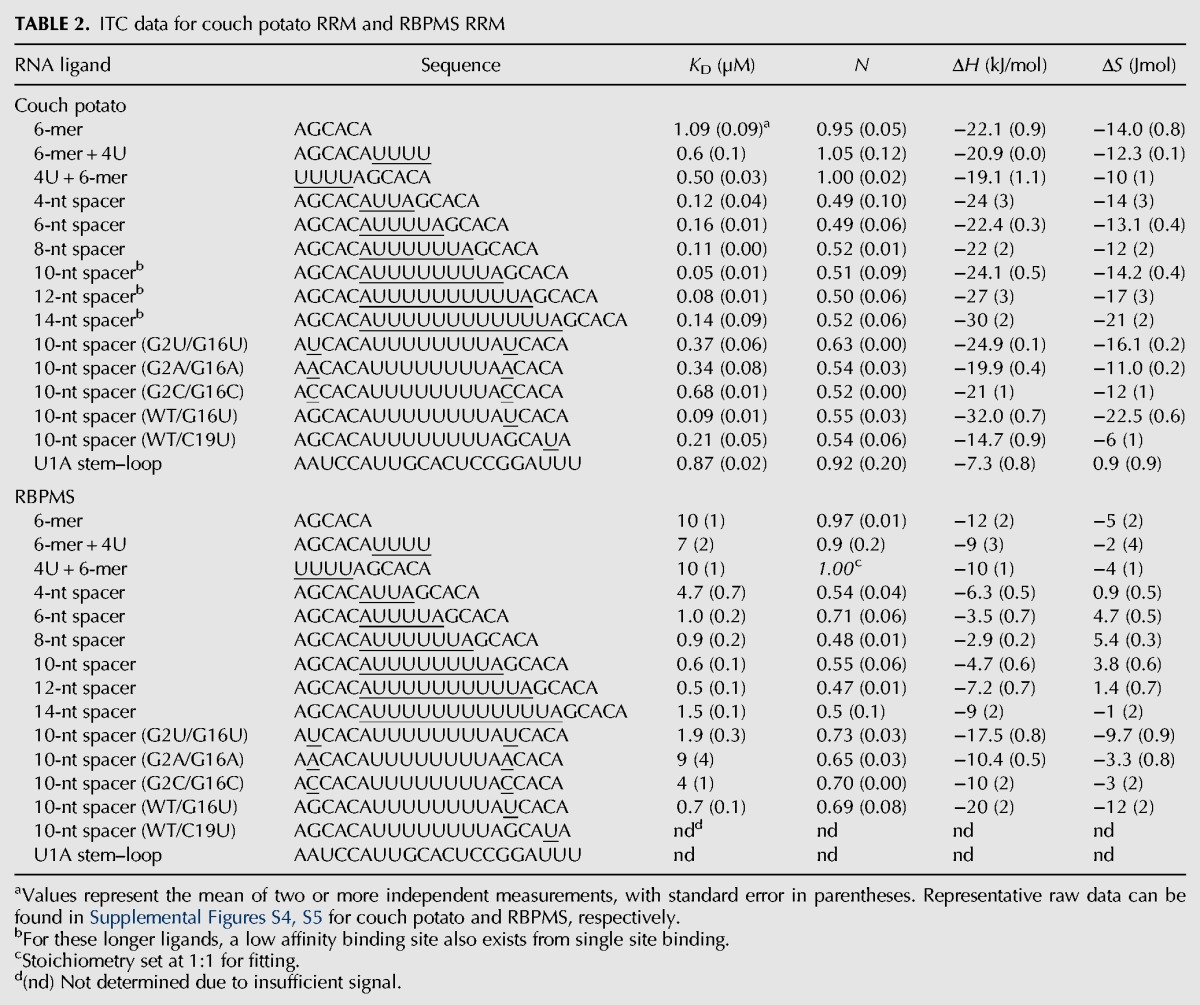
RNA binding preference for the guanine is also conserved among the three proteins. Similar to the situation with MEC-8, mutation of the two guanine bases to uracil (UCAC), adenosine (ACAC), or cytosine (CCAC) in the 10-nt linker RNA caused a reduction in binding by a factor of 7–14 for couch potato RRM, and by a factor of three to 15 for RBPMS RRM (Table 2). With respect to RBPMS/Hermes, evidence for in vivo importance of the guanine in a GCAC motif was previously suggested by the study of the 240-nt Xenopus oocyte mitochondrial cloud localization signal (MCLS). The MCLS contains six repeats of a UGCAC sequence and a mutation of all six to UUCAC prevents proper RNA localization of Xcat2 RNA to the germinal granules (Chang et al. 2004). The localization of nanos RNA to germinal granules is also dependent on the MCLS, and colocalization studies demonstrate that both nanos RNA and RBPMS/Hermes accumulate in similar clusters (Aguero et al. 2016). In addition, it was shown that intact UGCAC repeats of the MCLS are required for interaction with RBPMS/Hermes.
Finally, it had been previously noted that molecular similarities exist between RNA binding of CAC sequences by RBPMS and the first RRM of the U1A protein (Teplova et al. 2016). The additional contacts to the initial guanine increase this similarity. Structural details of U1A bound to the loop of hairpin II of U1 snRNA (Oubridge et al. 1994) or the U1A 3′UTR polyadenylation inhibition element (Allain et al. 1996, 1997; Varani et al. 2000) illustrate a similar RNA bound conformation for all four bases in the GCAC motif as compared to MEC-8. We therefore selected one of the U1 hairpin II RNA ligands (Fig. 2E; Oubridge et al. 1994) for assessment of binding to MEC-8 RRM1. Under the same conditions as the previous ITC measurements, this stem–loop RNA indeed bound with high affinity to MEC-8 RRM1, with a KD of 0.24 ± 0.02 µM, comparable to that of the tandem GCAC ligand (Table 1) although not as tight as the interaction between U1A and the stem–loop (Lutz-Freyermuth et al. 1990; Hall and Stump 1992). Binding to couch potato RRM was relatively weaker, but still with higher affinity than to the short AGCACA RNA (Table 2; Fig. 2E). The affinity to RBPMS could not be determined due to insufficient signal to noise. Nevertheless, it is evident that at least MEC-8 and couch potato from the RBPMS family are able to recognize the GCAC motif even within the context of a stem–loop structure.
In summary, we have used a combination of structure determination and binding assays to demonstrate the binding preference of MEC-8 RRM1 for GCAC motifs, and further show that optimal targets have two motifs separated by more than six bases. These results confirm the preferred GCAC motif identified for MEC-8 and couch potato by the RNA compete method (Ray et al. 2013), but differ slightly from the smaller CAC motif obtained from the in vivo assay for RBPMS targets by the PAR-CLIP approach (Farazi et al. 2014). For the in vivo targets, it is possible that additional protein partners in the cell could enhance binding affinity to less optimal sites, such as the interaction between Xenopus RBPMS/Hermes and hnRNP1 (Aguero et al. 2016). Nevertheless, some of the identified targets such as NDUFA6 and ETF1, which were further investigated by in vitro binding assays (Farazi et al. 2014; Teplova et al. 2016), do in fact fulfill the general observed requirement of at least one GCAC motif and a separation of >6 nt to a second suboptimal motif. The region of NOGGIN mRNA that interacts with RBPMS2 (nucleotides 570–730) (Sagnol et al. 2014) also contains two GCAC sequences separated by 18 bases that could serve as the RBPMS2 binding site. Extensive separation such as this could complicate analysis, as could the allowed presentation of the GCAC sequence within RNA secondary structure elements. Precise identification of in vivo sites bound by MEC-8 and couch potato will greatly add to the description of binding site determinants, and also eventually to functional understanding of the RBPMS family of RNA-binding proteins. Finally, it should be noted that we have focused on the only RRM domain that is absolutely conserved within the entire RBPMS family and additionally in all of the generated splice isoforms of RBPMS, MEC-8 and couch potato. Nevertheless, some family member isoforms (such as the MEC-8 variant shown in Fig. 1A) also possess a second C-terminal RRM domain with the ability to bind RNA. The resulting interplay regarding RNA-binding by the first and second RRM domains, and whether or not the different isoforms are specific to a particular biological process, represents a further complexity to be investigated.
MATERIALS AND METHODS
Protein expression and purification
The N-terminal RRM domain from C. elegans MEC-8 (residues 28–123 or 28–117) was amplified from a codon-optimized synthetic gene (Bio-Basic Canada) by using PCR oligos containing NcoI or Acc65I restriction enzyme sites, and inserted into a modified pET-9d plasmid encoding a His6 purification tag followed by a tobacco etch virus (TEV) protease cleavage site. To aid in sample stability, Cys54 and Cys102 were mutated to alanine. The orthologous RRM domains from human RBPMS (residues 11–111) and Drosophila couch potato (CPO; residues 447–543) were amplified from I.M.A.G.E. clone IRAUp969A1214D (BioValley) and EST clone BDGPp6122L092Q (BioValley), respectively, and inserted into the same modified pET-9d plasmid. MEC-8 mutant proteins were generated by using PCR amplification with a set of oligos overlapping the mutation site. All proteins were expressed in Escherichia coli BL21 LysY (New England Biolabs) using LB medium or M9 minimum medium supplemented with 15NH4Cl (1 g L−1) and [13C6]glucose (2 g L−1). Induction with 0.5 mM IPTG (final concentration) was followed by overnight protein expression at 20°C or 30°C for LB or M9 cultures, respectively. Bacteria were collected by centrifugation and resuspended in 50 mM Tris (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol, and 5 mM imidazole. Cell lysate preparation used a combination of lysozyme and sonication followed by centrifugation at 20,000g for 30 min. Protein purification from cell lysate was achieved by using Ni2+-affinity chromatography with buffers composed of 50 mM Tris (pH 7.5), 500 mM NaCl, 5% (v/v) glycerol, and 5, 25, and 250 mm imidazole for the loading, wash and elution, respectively. The His6 tag was removed by addition of TEV protease followed by a second Ni2+-affinity chromatography step. The purified samples were concentrated by filter centrifugation, dialyzed overnight, and quantified by measuring the absorbance at 280 nm. The RNA was synthesized by using an Expedite 8909 (PerSeptive Biosystems). The DNA oligonucleotides were commercially produced (Eurogentec and Eurofins MWG Operon).
NMR spectroscopy
Spectra were recorded at 298 K using a Bruker Avance III 700 MHz spectrometer equipped with a triple resonance-gradient standard probe. Topspin version 2.1 (Bruker BioSpin) was used for data collection. Spectra processing used NMRPipe (Delaglio et al. 1995) followed by analysis with Sparky 3 (T.D. Goddard and D.G. Kneller, University of California) or NMRviewJ 8.0 (One Moon Scientific). For backbone resonance assignment of 300 µM 13C,15N MEC-8 RRM1 the following spectra were used: 2D 1H-15N HSQC, 3D HNCO, 3D HNCA, 3D HNHA, 3D (H)CCONH TOCSY, and 3D H(C)CONH TOCSY. All spectra were collected in buffer containing 90% H2O/10% D2O. Amide 15N T1 and T2 relaxation data were acquired at 700 MHz and 298 K as previously described (Farrow et al. 1994). Steady-state heteronuclear {1H}15N-NOE spectra were recorded with and without 3 sec of 1H saturation. Relaxation rates and error calculations were determined using NMRViewJ (Johnson and Blevins 1994). Backbone 1H, 13C, and 15N chemical shift assignments for MEC-8 RRM1 were deposited in the Biological Magnetic Resonance Data Bank (BMRB) as entry 26922.
Crystallization and structure calculation
Purified recombinant MEC-8 RRM1 was concentrated to ∼10 mg/mL in 50 mM Tris (pH 7.5), 150 mM NaCl. Crystals of MEC-8 RRM1 (residues 23–117) were obtained by using 1 µL of the protein solution mixed with an equal volume of reservoir solution containing 0.1 M sodium acetate (pH 5.6), 0.2 M ammonium sulfate and 30% (w/v) polyethylene glycol (PEG) 4000 and using sitting drop vapor diffusion against 500 µL of reservoir solution. Crystals appeared as long plates. The ligand-bound complex was assembled by mixing the protein (10 mg/mL) with AGCACATTTTTTTTAGCACA DNA at a 2:1 molar ratio. X-shaped crystals were generated from a 2 µL 1:1 mixture of sample and reservoir buffer containing 10% (v/v) isopropanol and 10% (w/v) PEG4000 in 500 µL of reservoir. Crystal cryo-protection involved soaking for 5 min in a solution composed of 6.7% (v/v) isopropanol, 6.7% (v/v) PEG4000, and 25% (v/v) glycerol. Diffraction data were collected at the European Synchrotron Radiation Facility beam line ID23-2 and processed using the program XDS (Kabsch 2010). The unbound structure was determined by molecular replacement with Phaser (McCoy et al. 2007), based on the crystal structure of the second RRM domain of yeast poly(U)-binding protein PDB 3MD1 chain A. Molecular replacement for the DNA-bound complex used the unbound MEC-8 RRM1 dimer. Phenix was used for refinement (Adams et al. 2010). A summary of data collection and structure refinement statistics is provided in Supplemental Table S1. Structure images were prepared by using the program Chimera (Pettersen et al. 2004). Structural coordinates and data have been deposited in the Protein Data Bank as PDB ID codes 5BJR and 5TKZ for the unbound and ligand bound MEC-8 RRM1, respectively.
Isothermal titration calorimetry
ITC measurements were performed by using an ITC200 Microcal calorimeter (Malvern Instruments) at 25°C with a stir rate of 500 rpm and set at high sensitivity. All proteins were dialyzed extensively against 50 mM Tris, pH 7.5, 150 mM NaCl. Buffer from the dialysis was used to solubilize the DNA oligonucleotides, or used in conjunction with NAP-5 columns (GE Healthcare Life Sciences) to change the buffer of RNA samples. The initial determination of binding preference in Figure 1F represents an average of two measurements using 160–320 µM protein in the syringe and 20–40 µM ligands in the cell, with a total of 13 injections of 3 µL following an initial injection of 1 µL. For the sequence logo, minimal binding was set at a KA of 10000. Measurement of RNA binding by MEC-8 RRM1, couch potato RRM and RBPMS RRM (Tables 1, 2) used an average of at least two measurements. Samples contained 70–400 µM RNA in the syringe and 11–34 µM protein in the cell, depending on the signal and stoichiometry, with a total of 19 injections of 2 µL following an initial injection of 0.5 µL. The data were first processed by using NITPIC (Keller et al. 2012; Scheuermann and Brautigam 2015) and the integrated data points subsequently fit by using SEDPHAT (Zhao et al. 2015). The graph in Figure 1F was prepared by using GUSSI (Brautigam 2015).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gavin Collie for final preparation of crystals and synchrotron data collection. Additional assistance and advice were provided by Lionel Beaurepaire, Brice Kauffmann, Axel Innis, and Sébastien Fribourg. Brune Vialet provided synthesis of the RNA oligonucleotides. Access to NMR spectrometers, equipment, and technical assistance were possible thanks to Axelle Grélard, Estelle Morvan, and the structural biology platform at the Institut Européen de Chimie et Biologie (UMS 3033). We thank the European Synchrotron Radiation Facility for access to synchrotron radiation facilities (beam line ID23-2). Financial support from the Centre Nationale de la Recherche Scientifique (IR-RMN-THC Fr3050) is gratefully acknowledged. H.S. was supported by a PhD fellowship from the Institut National de la Santé et de la Recherche (INSERM) and the Aquitaine regional government.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059733.116.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguero T, Zhou Y, Kloc M, Chang P, Houliston E, Lou King M. 2016. Hermes (Rbpms) is a critical component of RNP complexes that sequester germline RNAs during oogenesis. J Dev Biol 4: 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Gubser CC, Howe PW, Nagai K, Neuhaus D, Varani G. 1996. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature 380: 646–650. [DOI] [PubMed] [Google Scholar]
- Allain FHT, Howe PWA, Neuhaus D, Varani G. 1997. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J 16: 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrane S, Rebora K, Zniber I, Dupuy D, Mackereth CD. 2014. Backbone-independent nucleic acid binding by splicing factor SUP-12 reveals key aspects of molecular recognition. Nat Commun 5: 4595. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Kooyer S, D'Evelyn D, Pearlman J. 1992a. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev 6: 2125–2136. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Vaessin H, Bier E, Kolodkin A, D'Evelyn D, Kooyer S, Jan YN. 1992b. The Drosophila couch potato gene: an essential gene required for normal adult behavior. Genetics 131: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA. 2015. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol 562: 109–133. [DOI] [PubMed] [Google Scholar]
- Calixto A, Ma C, Chalfie M. 2010. Conditional gene expression and RNAi using MEC-8-dependent splicing in C. elegans. Nat Methods 7: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. 1981. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol 82: 358–370. [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, Lou King M. 2004. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15: 4669–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daragan VA, Mayo KH. 1997. Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog Nucl Magn Reson Spectrosc 31: 63–105. [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Leonhardt CS, Mukherjee N, Mihailovic A, Li S, Max KE, Meyer C, Yamaji M, Cekan P, Jacobs NC, et al. 2014. Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets. RNA 20: 1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow NA, Zhang O, Forman-Kay JD, Kay LE. 1994. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR 4: 727–734. [DOI] [PubMed] [Google Scholar]
- Fu J, Cheng L, Wang Y, Yuan P, Xu X, Ding L, Zhang H, Jiang K, Song H, Chen Z, et al. 2015. The RNA-binding protein RBPMS1 represses AP-1 signaling and regulates breast cancer cell proliferation and migration. Biochim Biophys Acta 1853: 1–13. [DOI] [PubMed] [Google Scholar]
- Furukawa MT, Sakamoto H, Inoue K. 2015. Interaction and colocalization of HERMES/RBPMS with NonO, PSF, and G3BP1 in neuronal cytoplasmic RNP granules in mouse retinal line cells. Genes Cells 20: 257–266. [DOI] [PubMed] [Google Scholar]
- Gerber WV, Yatskievych TA, Antin PB, Correia KM, Conlon RA, Krieg PA. 1999. The RNA-binding protein gene, hermes, is expressed at high levels in the developing heart. Mech Dev 80: 77–86. [DOI] [PubMed] [Google Scholar]
- Gerber WV, Vokes SA, Zearfoss NR, Krieg PA. 2002. A role for the RNA-binding protein, hermes, in the regulation of heart development. Dev Biol 247: 116–126. [DOI] [PubMed] [Google Scholar]
- Glasscock E, Tanouye MA. 2005. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics 169: 2137–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB, Stump WT. 1992. Interaction of N-terminal domain of U1A protein with an RNA stem/loop. Nucleic Acids Res 20: 4283–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapkova I, Skarda J, Rouleau C, Thys A, Notarnicola C, Janikova M, Bernex F, Rypka M, Vanderwinden JM, Faure S, et al. 2013. High expression of the RNA-binding protein RBPMS2 in gastrointestinal stromal tumors. Exp Mol Pathol 94: 314–321. [DOI] [PubMed] [Google Scholar]
- Hornberg H, Wollerton-van Horck F, Maurus D, Zwart M, Svoboda H, Harris WA, Holt CE. 2013. RNA-binding protein Hermes/RBPMS inversely affects synapse density and axon arbor formation in retinal ganglion cells in vivo. J Neurosci 33: 10384–10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. 1994. NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. 2012. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem 84: 5066–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong JMK, Caprioli J, Piri N. 2010. RNA binding protein with multiple splicing: A new marker for retinal ganglion cells. Investig Ophthalmol Vis Sci 51: 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Herman RK. 1994. The mec-8 gene of Caenorhabditis elegans affects muscle and sensory neuron function and interacts with three other genes: unc-52, smu-1 and smu-2. Genetics 138: 83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, Shaw JE. 1996. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122: 1601–1610. [DOI] [PubMed] [Google Scholar]
- Lutz-Freyermuth C, Query CC, Keene JD. 1990. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc Natl Acad Sci 87: 6393–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarnicola C, Rouleau C, Le Guen L, Virsolvy A, Richard S, Faure S, De Santa Barbara P. 2012. The RNA-binding protein RBPMS2 regulates development of gastrointestinal smooth muscle. Gastroenterology 143: 687–697. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Piri N, Kwong JMK, Song M, Caprioli J. 2006. Expression of hermes gene is restricted to the ganglion cells in the retina. Neurosci Lett 405: 40–45. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AR, de Sevilla Müller LP, Brecha NC. 2014. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol 522: 1411–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagnol S, Yang Y, Bessin Y, Allemand F, Hapkova I, Notarnicola C, Guichou J-F, Faure S, Labesse G, de Santa Barbara P. 2014. Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity. Nucleic Acids Res 42: 10173–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen TS, Vesala L, Laiho A, Merisalo M, Hoikkala A, Kankare M. 2015. Seasonal gene expression kinetics between diapause phases in Drosophila virilis group species and overwintering differences between diapausing and non-diapausing females. Sci Rep 5: 11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann TH, Brautigam CA. 2015. High-precision, automated integration of multiple isothermal titration calorimetric thermograms: new features of NITPIC. Methods 76: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Zhu C-T, Das J, Batavia M, Yang L, Eanes WF. 2008. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci 105: 16207–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Kitao S, Ichikawa K, Suzuki N, Yamabe Y, Imamura O, Tokutake Y, Satoh M, Matsumoto T, Kuromitsu J, et al. 1996. A unique human gene that spans over 230 kb in the human chromosome 8p11-12 and codes multiple family proteins sharing RNA-binding motifs. Proc Natl Acad Sci 93: 10913–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, Cauffman K, Chan AP, Zhou Y, Lou King M, Etkin LD, Kloc M. 2007. Hermes RNA-binding protein targets RNAs-encoding proteins involved in meiotic maturation, early cleavage, and germline development. Differentiation 75: 519–528. [DOI] [PubMed] [Google Scholar]
- Spike CA, Davies AG, Shaw JE, Herman RK. 2002. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129: 4999–5008. [DOI] [PubMed] [Google Scholar]
- Teplova M, Farazi TA, Tuschl T, Patel DJ. 2016. Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS. Q Rev Biophys 49: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Gunderson SI, Mattaj IW, Kay LE, Neuhaus D, Varani G. 2000. The NMR structure of the 38 kDa U1A protein—PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol 7: 329–335. [DOI] [PubMed] [Google Scholar]
- Wilmore HP, McClive PJ, Smith CA, Sinclair AH. 2005. Expression profile of the RNA-binding protein gene hermes during chicken embryonic development. Dev Dyn 233: 1045–1051. [DOI] [PubMed] [Google Scholar]
- Zearfoss NR, Chan AP, Wu CF, Kloc M, Etkin LD. 2004. Hermes is a localized factor regulating cleavage of vegetal blastomeres in Xenopus laevis. Dev Biol 267: 60–71. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piszczek G, Schuck P. 2015. SEDPHAT–a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.