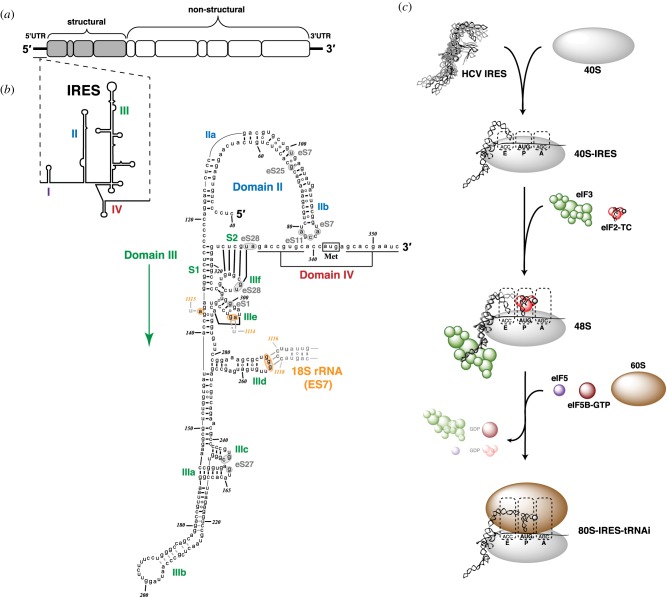
Figure 4.
HCV IRES secondary structure and mechanism. (a) Schematic of the HCV positive-sense RNA genome. (b) Secondary structure, domain architecture and ribosomal interactions of HCV IRES RNA. Intra-IRES and IRES : rRNA base-pairing is based on conformation of the ribosome-bound IRES [71,72]. Base-pairing of nt 178–221 of domain IIIb are drawn in grey as they are not resolved in the 40S : IRES structure, and are thus hypothetical. The short hairpin of domain IV is shown unfolded, encompassing the ribosomal toeprint-defined region of the 40S : IRES complex [73]. (c) Model of HCV IRES-mediated initiation: 40S subunit recruitment to form the binary complex, initiator tRNA recruitment during 48S PIC formation and 60S subunit joining to form the elongation-competent 80S ribosome.