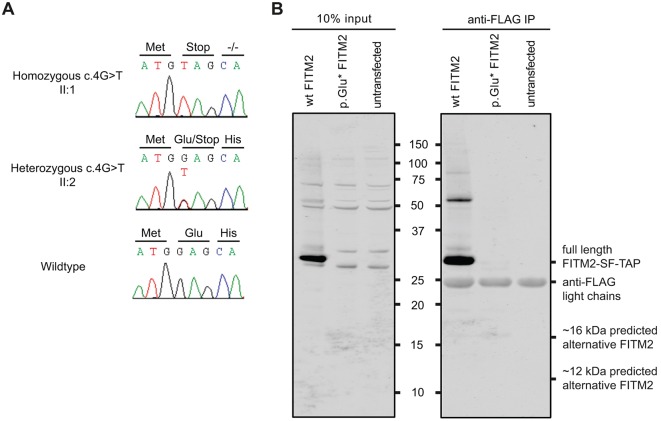
Fig. 2.
Identification of a genetic defect underlying syndromic hearing impairment in family W09-1008 and expression analysis of wild-type and p.Glu2* FITM2 fused to a FLAG-tag in HEK293T cells. (A) Partial sequences of FITM2 exon 1 are shown from an affected member, an unaffected heterozygous sib and an unaffected wild-type sib of family W09-1008. The predicted amino acid changes and the surrounding amino acids are indicated above the sequence. Sequence NM_001080472.1 was employed as a reference. (B) The left panel shows a western blot of a gel on which 10% of the cell lysate was loaded (before affinity purification). The right panel shows a western blot of a gel on which 10 µl of the lysate after affinity purification (anti-FLAG) was loaded. Wild-type (WT) FITM2 migrates around 29 kDa and it is absent upon transfection and expression of the p.Glu2* FITM2 construct. After affinity purification, a very weak band is observed at ∼16 kDa. However, the intensity of the 16 kDa band is about 2700-fold lower than the wild-type FITM2 band and therefore it is likely to have little or no biological impact. The gel was immunostained with an anti-FLAG polyclonal antibody. Of four ATG-triplets in the original reading frame, three (codon positions 94, 493 and 508) are predicted to be potential translation initiation sites by the Netstart 1.0 algorithm (Pedersen et al., 1997). Accordingly, alternative proteins would consist of 285 amino acids (aa) (26.2 kDa), 152 aa (11.2 kDa), and 147 aa (10.6 kDa), respectively. Expression constructs encode FITM2 fused to a C-terminal Strep-FLAG-tag (SF-TAP), adding approximately 6 kDa to the proteins. Wild-type FITM2 was found to migrate according to molecular weight of ∼29 kDa, which is lower than the predicted mass of the complete protein. However, fragments of similar length are observed with anti-FITM2 staining in the work of Duckert et al. (2004) and pro-peptide cleavage is predicted by the ProP algorithm. Marker size is indicated between the panels and given in kDa.