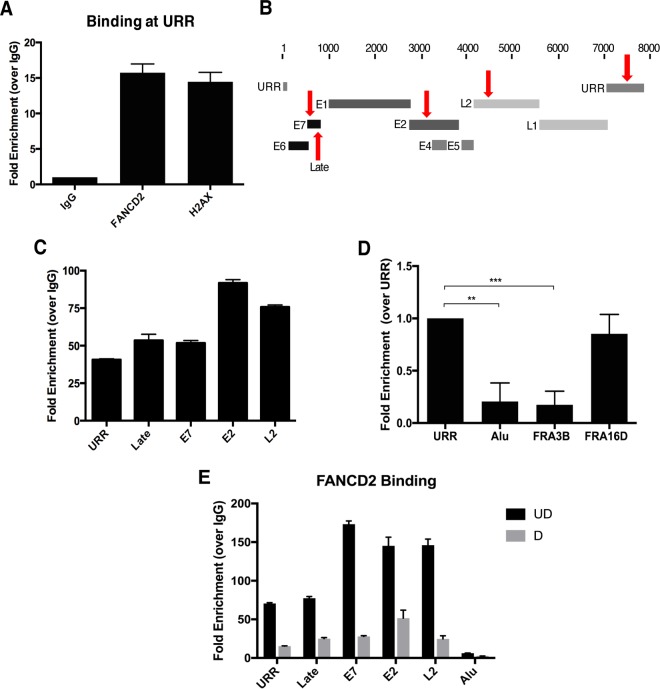
FIG 6 .
FANCD2 is preferentially recruited to HPV DNA. (A) Chromatin immunoprecipitation (ChIP) analysis of FANCD2 and γH2AX binding to the URR in CIN612 cells. Quantitative real-time PCR (qRT-PCR) was performed using a LightCycler 480 (Roche), and fold enrichment was quantitated relative to an IgG control. Similar results were seen in three independent experiments. Error bars represent the standard deviations between experiments. (B) Schematic of the HPV31 linearized genome, with primer regions indicated with arrows. (C) ChIP analysis for FANCD2 binding at indicated sites in the viral genome. Fold enrichment was normalized to an IgG control. Similar results were seen in three independent experiments. Error bars represent the standard deviations between experiments. (D) ChIP analysis of FANCD2 binding at the URR compared to Alu repeat and fragile site regions (FRA3B and FRA16D) in the host genome. Enrichment was normalized to an IgG control and is represented as fold change over URR across three independent experiments. The graph represented as percentage of input shows a similar trend (Fig. S1). Error bars represent the standard deviations between experiments. A standard Student’s t test was used to determine statistical significance. **, P < 0.005; ***, P < 0.0005. (E) CIN612 cells were differentiated for 72 h in 1.5 mM calcium medium, and ChIP analysis was performed for binding across the HPV genome. Fold enrichment was normalized to an IgG control. Similar results were seen in three independent experiments. Error bars represent the standard deviations between experiments. UD, undifferentiated; D, differentiated.