SUMMARY
Eukaryotic genomes are packaged in chromatin. The higher-order organization of nucleosome core particles is controlled by the association of the intervening linker DNA with either the linker histone H1 or high mobility group box (HMGB) proteins. While H1 is thought to stabilize the nucleosome by preventing DNA unwrapping, the DNA bending imposed by HMGB may propagate to the nucleosome to destabilize chromatin. For metazoan H1, chromatin compaction requires its lysine-rich C-terminal domain, a domain that is buried between globular domains in the previously characterized yeast Saccharomyces cerevisiae linker histone Hho1p. Here, we discuss the functions of S. cerevisiae HMO1, an HMGB family protein unique in containing a terminal lysine-rich domain and in stabilizing genomic DNA. On ribosomal DNA (rDNA) and genes encoding ribosomal proteins, HMO1 appears to exert its role primarily by stabilizing nucleosome-free regions or “fragile” nucleosomes. During replication, HMO1 likewise appears to ensure low nucleosome density at DNA junctions associated with the DNA damage response or the need for topoisomerases to resolve catenanes. Notably, HMO1 shares with the mammalian linker histone H1 the ability to stabilize chromatin, as evidenced by the absence of HMO1 creating a more dynamic chromatin environment that is more sensitive to nuclease digestion and in which chromatin-remodeling events associated with DNA double-strand break repair occur faster; such chromatin stabilization requires the lysine-rich extension of HMO1. Thus, HMO1 appears to have evolved a unique linker histone-like function involving the ability to stabilize both conventional nucleosome arrays as well as DNA regions characterized by low nucleosome density or the presence of noncanonical nucleosomes.
KEYWORDS: HMGB proteins, linker histone, Saccharomyces cerevisiae, chromatin, nucleosome
INTRODUCTION
The DNA of all eukaryotic cells is tightly packaged into chromatin, a nucleoprotein complex consisting of DNA associated with histone and nonhistone proteins. The nucleosome is the fundamental unit of chromatin in which ∼146 bp of DNA wrap around the histone octamer composed of two copies each of H2A, H2B, H3, and H4. The linker DNA connecting nucleosome core particles may associate with either linker histone H1 or nonhistone proteins such as the high mobility group box (HMGB) proteins, which often act in opposition to H1. In this review, we discuss the Saccharomyces cerevisiae HMGB protein HMO1, which is unique in encompassing features of both canonical HMGB proteins and linker histones. We consider its role in stabilizing nucleosome-free DNA and in modulating chromatin stability and dynamics during DNA repair and transcription.
Linker Histones
Nucleosome structure and organization.
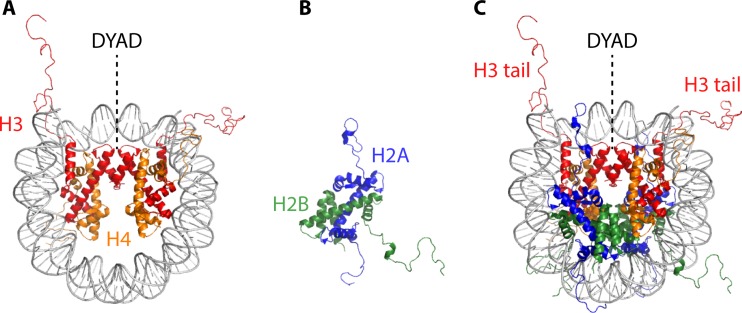
The static structure of the nucleosome core particle has been determined at high resolution, and the folding of a four-nucleosome array has also been reported (1, 2). The nucleosome core particle consists of a histone octamer composed of two H2A/H2B heterodimers and an (H3/H4)2 heterotetramer about which ∼146 bp of DNA is wrapped ∼1.7 times in a left-handed supercoil (Fig. 1). Thanks to the identification of flexible DNA sequences that preferentially associate with core histones, it has been possible to achieve high-resolution structural information (3, 4). In vitro, nucleosome formation at a specific sequence is directed by intrinsic properties of the DNA, and it is nucleated by the association of the (H3/H4)2 tetramer, which marks the initial point of DNA bending and therefore defines the dyad axis (Fig. 1A); the binding of (H3/H4)2 is followed by the deposition of two H2A/H2B dimers (5–7). In vivo, nucleosome assembly is catalyzed by chaperones (8–10). The histone octamer makes numerous direct contacts to DNA, mostly in the DNA minor grooves, and the resulting DNA structure deviates significantly from the canonical B-form. Adjacent nucleosomes are connected by linker DNAs of various lengths to generate nucleosomal arrays (Fig. 2); the N-terminal tails of core histones extend away from the nucleosome core particle, and these positively charged extensions have been implicated in contacts to DNA and to neighboring nucleosomes.
FIG 1.
Assembly of the nucleosome core particle. (A) Association of the (H3/H4)2 tetramer with DNA nucleates nucleosome assembly and defines the dyad axis. (B) H2A/H2B dimer. (C) Two H2A/H2B dimers are deposited to generate the nucleosome core particle. H3 N-terminal tails emerge near the DNA entry/exit points (based on data reported under PDB accession number 1KX5).
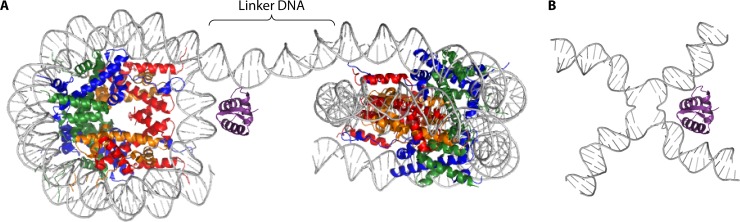
FIG 2.
Histone H1 associates with linker DNA. (A) The globular domain of histone H1 (purple) binds the nucleosome at the dyad. The structure of a dinucleosome is depicted; the color coding for core histones is the same as that in Fig. 1. (B) Four-way junction DNA mimics the DNA configuration at the nucleosome dyad, perhaps explaining the preferred binding of H1 to such junctions. The dinucleosome represents the asymmetric unit in the structure of a tetranucleosome with one linker DNA trimmed for clarity (PDB accession number 1ZBB) (2). The H1 globular domain and its localization relative to the dyad are based on the structure of the chicken H5 globular domain in complex with a nucleosome (PDB accession number 4QLC) (33). The representation of the four-way junction is based on data reported under PDB accession number 3CRX.
Higher-order levels of organization in which nucleosomal arrays associate with other proteins remain poorly understood. Interactions between nucleosomes promote the folding of the nucleosomal array into a more compact 30-nm fiber, for example, by the interaction of the H4 N-terminal tail with an acidic patch formed at the H2A/H2B interface on a neighboring nucleosome (11). The linker histone H1 plays an indispensable role in stabilizing the 30-nm fiber in which nucleosomes are clustered tightly together, decreasing the internucleosomal distance and fixing the entry/exit angle of DNA (12–16). This compaction is affected by the nucleosomal repeat length, as the repeat length must be sufficient to accommodate H1 binding; for nucleosomal arrays with shorter repeat lengths, internucleosome interactions drive the folding of a more compact fiber that is less affected by linker histone binding (17). However, in proliferating cells, evidence of 30-nm fibers is lacking, and chromosome organization is instead thought to involve a zigzag geometry and long-range looping that is modulated by the density of linker histones (18–22). This organization is thought to involve the formation of topologically associated domains by the formation of loops within higher-order chromatin structures; precisely how H1 mechanistically participates is unresolved.
Linker histone binding to the nucleosome.
Histone H1 binds linker DNA where it enters and exits the nucleosome (Fig. 2) (23–26). Unlike core histones, which have residency times on a time scale of hours, linker histones are quite mobile, with residency times being measured in minutes (16, 27, 28). In mammalian cells, H1 is present at an average stoichiometry of ∼0.8 molecules per nucleosome, with variations depending on the cell type, and in chicken, the ratio of linker histones to nucleosomes was estimated to be 1.3 histones per nucleosome (29). Metazoan linker histones have a tripartite structure. They interact with about 20 bp of DNA (either asymmetrically by preferentially binding one linker segment or by protecting 10 bp of entering and exiting DNA) to create the chromatosome, consisting of ∼167 bp of DNA, the core histone octamer, and one molecule of H1. The ∼40-amino-acid N-terminal domain is followed by the highly conserved central globular domain of ∼80 amino acids and a long C-terminal domain (CTD) (∼100 amino acids) (Fig. 3) (30). The globular domain adopts a winged-helix DNA binding motif (31); its interaction with DNA at the nucleosomal entry/exit points gives rise to protection of the additional ∼20 bp (23, 32). The structure of the chicken linker histone H5 in complex with a nucleosome reveals the binding of the globular domain on the nucleosome dyad axis, interacting with both DNA linkers, whereas the Drosophila melanogaster linker histone H1 binds off-dyad (32, 33). Experiments involving site-directed mutagenesis followed by determination of binding to nucleosomes in living cells have likewise led to the conclusion that the globular domains of different mouse H1 isoforms use different interaction surfaces for contacts to chromatin (34, 35). This suggests that interactions with linker histones in different binding modes might differentially control higher-order chromatin organization. In general, linker histones bind preferentially to four-way DNA junctions compared to linear DNA (36), and binding to the nucleosome at the DNA entry/exit points is thought to reflect this preference for a specific DNA geometry (Fig. 2).
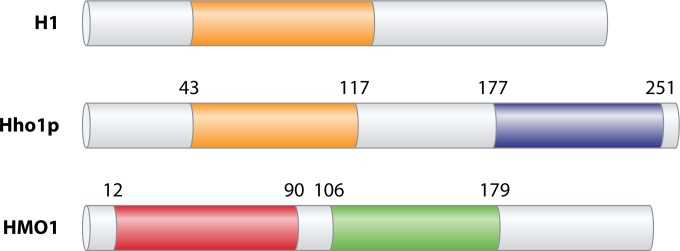
FIG 3.
Domain organization of H1, Hho1p, and HMO1. Metazoan H1 typically contains an ∼40-amino-acid N-terminal extension, followed by a globular domain of ∼80 amino acids (orange) and a long CTD characterized by S/TPXK-like repeats. Hho1p contains a lysine-rich N-terminal segment followed by a globular domain with similarity to that of H1 (orange). Another lysine-rich segment connects this globular domain to the second globular domain (purple). HMO1 contains box A (red), which has little similarity to consensus HMG domains, followed by a lysine-rich linker, the box B domain (green), and a lysine-rich CTD. Mammalian HMGB proteins have a domain organization similar to that of HMO1, except that the CTD is acidic.
Notably, the regions flanking the globular domain, particularly the lysine-rich CTD, are required for the formation of higher-order structures (Fig. 4) (37, 38). The low-complexity sequence of the CTD, which includes ∼40% lysine and a significant content of alanine and proline, results in the domain remaining unstructured in aqueous solution due to charge repulsion but acquiring a kinked-helix conformation when bound to DNA (39, 40). Interactions with the CTD promote the formation of higher-order chromatin structures as well as increase the residence time (23, 37, 41). Modeling suggests that a highly charged CTD compacts chromatin more effectively, resulting in silencing, whereas less-charged CTDs promote a chromatin folding in which the genome is more accessible (42). The N terminus, which is also unstructured, affects positioning and DNA binding affinity (43, 44). While specific H1 binding modes may be distinct for different H1 isoforms, current data support a general mode of binding in which the H1 globular domain binds near the dyad axis, with the CTD mainly contacting one linker DNA such that linker DNA is organized into a stem-like structure (Fig. 4) (32, 38). In this configuration, one H1 has been proposed to link three nucleosomes and to prevent the association of additional H1 protomers, likely due to electrostatic repulsion.
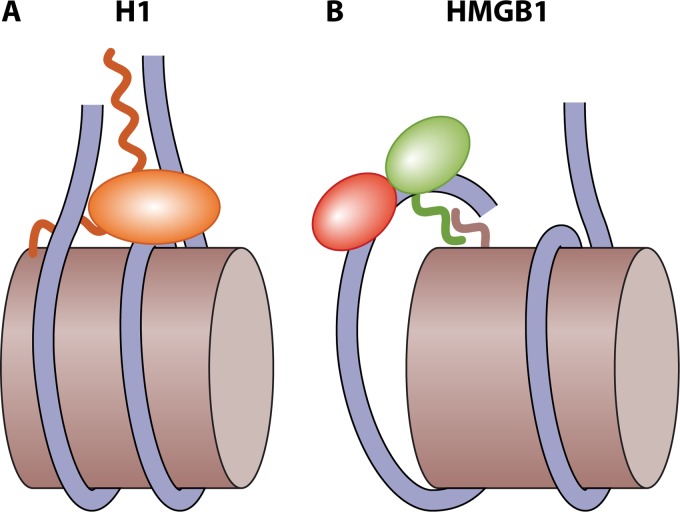
FIG 4.
Proposed interaction of H1 and HMGB proteins with nucleosomes. (A) H1 binds near the dyad such that the CTD mainly contacts one linker segment; this creates a stem-like structure that stabilizes the nucleosome core (32, 38). (B) For HMGB, interactions between the acidic CTD and the H3 N-terminal tail that exits near the dyad promote binding of HMGB to DNA; the DNA bending and underwinding induced may propagate to the nucleosome core to promote unwrapping or access to other factors (118, 119).
Diversity of linker histones.
The H1 family of linker histones is the most divergent class of histone proteins (45). For example, while the sequence of core histone H4 is 92% identical between yeasts and humans, the level of sequence identity between human H1 and the yeast linker histone Hho1p is only 31%. In addition, there are multiple different H1 subtypes in most eukaryotes (46). Some are constitutively expressed in all cells, while others are developmentally regulated, restricted to specific cell types, or induced at certain stages of differentiation. Covalent modifications contribute further to functional diversity (14–16, 47). Although the sequence of the winged-helix motif is relatively well conserved, the CTDs are extremely variable in both length and amino acid composition. Considering the role of the CTD in folding of nucleosomal arrays, different H1 isoforms are likely to exert different effects on chromatin organization. That the lysine-rich CTD is key to the organization of genomic DNA is reflected in euglenozoan protists, such as the kinetoplastids, which possess small linker histones that lack the winged-helix motif entirely and are similar to the basic CTD of metazoan histone H1 (48), although the amino acid composition may differ from that of the metazoan proteins. Such single-domain H1 proteins likely compact DNA by mechanisms that are distinct from those employed by metazoan H1. In contrast, the Gallus gallus (chicken) erythrocyte linker histone H5 shares greater sequence homology to human histone H1.0 (66% identity in comparisons of the complete sequences), with the CTD being more divergent (49).
Yeast linker histone Hho1p.
In contrast to higher eukaryotes, less is known about linker histone function in S. cerevisiae. Sequencing of the yeast genome showed the existence of an unusual linker histone H1, named Hho1p, characterized by having two globular domains, one of which has significant homology to the globular domain of metazoan H1 (Fig. 3) (50). A short basic tail precedes the H1-like globular domain, and the second globular domain follows a lysine-rich linker. No other linker histones that contain two globular domains have been reported. While the first globular domain closely resembles the winged-helix-turn-helix motif characteristic of metazoan H1, the second globular domain is unstructured under physiological conditions but adopts a winged-helix fold in the presence of high concentrations of tetrahedral anions (51). Only the first globular domain can associate with nucleosomes to protect additional DNA from nuclease digestion in vitro, whereas the second domain exhibits the greatest affinity for four-way junction DNA (52, 53). Four-way junction DNA may mimic the DNA conformation at nucleosomal entry/exit points (Fig. 2), and the ability of Hho1p to bind two four-way junction structures simultaneously has been reported, raising the possibility that Hho1p may bridge two adjacent nucleosomes (54); however, direct evidence for such binding has not been demonstrated.
Micrococcal nuclease (MNase) digests accessible linker DNA to produce DNA fragments corresponding to the chromatosome, whereas the digestion of nucleosomal arrays depleted of H1 is faster and generates shorter ∼146-bp fragments corresponding to the nucleosome core particle (55). In contrast to the nuclease sensitivity that results from eliminating H1, the absence of Hho1p does not result in a significant reorganization of nucleosomes or a change in the chromatin structure during vegetative growth, perhaps due to the absence of a terminal lysine-rich domain (50, 56). In addition, the stoichiometry of Hho1p to nucleosomes is lower than that of metazoan H1; estimates of cellular concentrations of Hho1p suggested a ratio of 1 Hho1p molecule per 37 nucleosomes (or ∼2,000 molecules/haploid cell) (57), a number that is in good agreement with a more recent estimate of the cellular protein content (∼2,600 molecules/cell) using a proteomics approach (58). A separate study indicated a ratio of 1 Hho1p molecule per 4 nucleosomes by comparison to the cellular content of histone H2A (59); however, since core histones are produced in excess over the quantities required for nucleosome assembly during S phase (60), this number may be overestimated. The proposed binding mode for Hho1p, in which its globular domains simultaneously engage adjacent nucleosomes, would be expected to generate a different type of nucleosome compaction from that of H1; in comparison, one H1 molecule has been proposed to link three nucleosomes to generate a zigzag pattern. Different binding modes would be consistent with differential sensitivities to MNase of H1- or Hho1p-containing chromatin (38, 54).
Genome-wide, Hho1p binding was shown to be variable and to be concentrated at ribosomal DNA (rDNA), where it has been implicated in repressing the expression of RNA polymerase II (Pol II)-transcribed reporter genes embedded in the rDNA, suggesting a role in rDNA compaction (61). A general role for Hho1p in the formation of DNA loops and for DNA compaction during stationary phase was also reported (62, 63). In contrast, Hho1p has also been demonstrated to prevent the establishment of silent chromatin, perhaps by modifying the barriers that separate transcriptionally active chromatin from heterochromatin (57, 63–65). Thus, both H1 and Hho1p have been implicated in long-range DNA looping and DNA compaction; however, the molecular mechanisms by which these proteins exert such functions are likely to differ.
Chromatin represses transcription.
In general, transcriptional repression correlates with chromatin condensation. Even nucleosomal arrays are repressive to transcription, as nucleosomes prevent transcription factors from accessing their cognate DNA. Promoters are therefore typically depleted of nucleosomes compared to the transcribed regions. Such nucleosome-free regions are found just upstream of the transcriptional start site, while the +1 nucleosome, which is found downstream of the start site, is localized strongly to this position. In yeast, nucleosome-free regions are typically maintained by transcription factors (66, 67).
Genome-wide profiling has demonstrated not only an absence of nucleosomes from active gene promoters in human cells but also a more extensive absence of linker histones, both upstream and downstream of the transcriptional start site (68). An early instructive example of transcriptional repression by H1 in Xenopus laevis shows that reduced H1 expression leads to the upregulation of 5S rRNA expression (69). More recent studies have suggested gene-specific transcriptional regulation by H1 as opposed to global effects and that H1 subtypes have an uneven distribution across the genome (70, 71). For example, chromatin immunoprecipitation (ChIP) showed a relative depletion of human H1.2 and H1.4 in actively transcribed chromatin, whereas all somatic subtypes were detected in heterochromatin (72). Consistent with this observation, H1.2 was reported to be overexpressed in cancer cells, where it is recruited to target genes by association with trimethylated H3 lysine 27 (H3K27me3) and contributes to the establishment of silent chromatin by a mechanism that requires its CTD (73). In heterochromatin, methylated H1 is implicated in the recruitment of factors such as heterochromatin protein 1 (HP1) (74, 75). Consistent with the ability of H1 to organize linker DNA into a stem-like structure (Fig. 4), H1 has been proposed to repress transcription by limiting nucleosome unwrapping as opposed to physically blocking transcription factor binding (76). This is consistent with genome-wide analyses that point to extensive H1 displacement in transcriptionally active genes (68, 77). Such a displacement may be aided by chaperones (78).
Chromatin represses DNA repair.
Genome integrity is continuously challenged by both endogenous and environmental agents that induce DNA damage. Such damage occurs in the context of chromatin, and higher-order chromatin structures are generally repressive for DNA repair. The “access-repair-restore” model describes sequential events involved in DNA repair in terms of detection of the lesion, chromatin remodeling to allow access to the repair machinery, the actual repair event, and, finally, restoration of the original chromatin state (79, 80). In this scenario, chromatin is viewed as a barrier that needs to be dismantled for DNA repair to proceed. However, the picture is more complex, and emerging evidence has pointed to a role for the nucleosome in recruiting DNA repair proteins (81).
Among the various DNA lesions, double-strand breaks (DSBs) are particularly genotoxic, and continuous DNA damage without efficient DSB repair may result in tumorigenesis and ageing. The primary DSB repair pathways include homologous recombination (HR) and nonhomologous end joining (NHEJ). Homologous recombination relies on DNA homology between sister chromatids and precisely repairs DSBs, while NHEJ is a more error-prone process that uses no or very limited sequence homology to rejoin two DNA ends (82). DNA repair proteins such as Ku and Rad51p play a critical role in DSB repair. Rad51p promotes homologous recombination to repair DSB lesions; however, the chromatosome inhibits homologous pairing. To overcome this barrier and to aid Rad51p-mediated homologous pairing, Rad54p, a member of the ATP-dependent nucleosome remodeling factor family, is required in both yeast and metazoans (83, 84). The linker histone functions as a negative regulator to suppress inappropriate DNA recombination, which may cause chromosomal aberrations. In S. cerevisiae, Hho1p was also reported to suppress homologous recombination (59, 85). To efficiently repair DSBs by Rad51p- and Rad54p-mediated homologous recombination, the linker histone H1 is evicted by a histone chaperone, Nap1p, from a reconstituted nucleosome array, suggesting that eviction of H1 promotes repair by homologous recombination (86). In contrast, Ku, which is integral to DSB repair by nonhomologous end joining, has been reported to readily displace human H1 from DNA ends (87). In human cells, ubiquitylation of H1 at DSB sites was also recently implicated in the recruitment of repair factors, adding to the collection of histone marks that contribute to repair events (88).
High Mobility Group Proteins
In eukaryotes, high mobility group (HMG) proteins are abundant nuclear proteins that make up a significant fraction of DNA binding nonhistone proteins. The HMGB protein family is the largest family of HMG proteins and a major nucleosome-binding constituent of the metazoan nucleus (89). In mammalian cells, the nucleus contains approximately 1 HMGB protein for every 10 nucleosomes (90). In addition to roles in DNA-dependent events, HMGB proteins sense cellular stress and function as extracellular cytokines, contributing to inflammatory and immune responses (91).
DNA binding by HMGB proteins.
The HMGB subfamily is divided into two classes, sequence-specific transcription factors that are expressed in a few cells and non-sequence-specific chromatin-associated proteins, which are abundant constituents of all eukaryotic nuclei. Transcription factors such as LEF-1 and SRY usually contain a single 80-amino-acid HMG box in which three α-helices create an L-shaped motif (Fig. 5B) (92). Most non-sequence-specific chromatin-associated HMGB proteins, e.g., mammalian HMGB1 to -4, possess two HMGB domains and bind preferentially to non-B-form DNA structures such as four-way junctions and DNA modified by the anticancer agent cisplatin (93, 94). Exceptions have been described, for example, in S. cerevisiae and Drosophila melanogaster, which encode single HMG box proteins (NHP6A/B and HMGD, respectively) that bind DNA without a sequence preference (95, 96).
FIG 5.
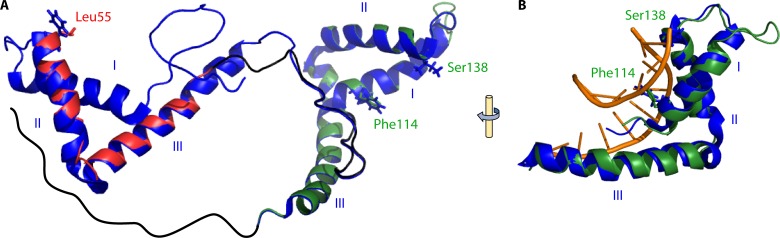
Model of HMO1 and its interaction with DNA. (A) HMO1 was modeled by using SwissModel in the automated mode, using human HMGB1 (PDB accession number 2YRQ) as the template. HMGB1 is shown in blue, and the HMO1 model is overlaid with box A and box B domains in red and green, respectively. Predicted HMO1-intercalating residues Leu55, from box A, and Phe114, from box B, are shown in a stick representation. Ser138 is located in the position occupied by the DNA-intercalating Ile residue in HMGB1 box B. Helices are identified with roman numerals. The HMO1 C-terminal extension (black) is inferred to interact with box A. (B) HMGB1 box B (blue) overlaid with HMO1 box B (green), showing the interaction of helix III in the DNA minor groove and the intercalation of Phe between DNA bases. HMGB1 box B DNA is based on data reported under PDB accession number 2GZK.
The HMG box serves as the primary site of binding to DNA and chromatin. The interaction of HMGB proteins with DNA is very dynamic; HMGB proteins bind transiently to B-form DNA and bend their DNA targets, and the mode of interaction of HMGB proteins with chromatin has therefore been characterized as a “hit and run” (93, 97). The energy required for DNA bending is derived from the extensive contacts of HMG boxes with the minor groove of DNA. Since the energetic cost of bending DNA is decreased in distorted or prebent DNA, HMGB1 proteins associate with such distorted DNA structures with high affinity (93, 98–100). The functional consequences of HMGB1 binding to damaged DNA have been alternately suggested to be a shielding of the lesion from the repair machinery or enhanced recognition of the damaged site (99, 101, 102). For example, HMGB1 has been reported to sensitize cells to cisplatin by impeding repair, perhaps influenced by the cellular redox state (103, 104); conversely, human HMGB1 has been reported to facilitate nucleotide excision repair (NER) by recruiting the NER protein XPA to interstrand cross-links (105). Interestingly, the recruitment of XPA to nondamaged sites was increased in HMGB1-depleted cells, suggesting that HMGB1 not only promotes NER but also facilitates the specificity of XPA-mediated damage recognition.
Structure of HMGB proteins.
The canonical HMGB proteins have a molecular mass of ∼25 kDa, containing two similar HMG domains, boxes A and B, and a C-terminal tail of ∼30 acidic amino acids (Fig. 3). Despite their similarity, box A differs from box B in the relative orientations of helices I and II and in the trajectory of the helix I-helix II loop, and the two domains have distinct electrostatic surface potentials in their DNA binding regions. The concave sides of both domains bind in the minor groove of DNA by using van der Waals and electrostatic interactions to induce a bend toward the major groove. The A domain has a greater preference for distorted DNA (106, 107), whereas the B domain binds less selectively to distorted DNA structures but can introduce an approximately right-angled bend into linear DNA (108). Partial DNA intercalation of hydrophobic residues located toward the N terminus of helices I and II introduces a kink into the bound DNA and thus enhances the bend associated with a widening of the minor groove (Fig. 5B). The bends induced by either domain likely reinforce each other (109, 110). For HMGB1, acetylation of lysine residues in the box A domain occurs in vivo, and it has been reported that substitution of these lysine residues compromises the preferred binding to both four-way junction DNA and constrained minicircles (107).
An important feature of HMGB1 and HMGB2 is the presence of a long acidic C-terminal “tail” consisting of ∼30 acidic residues (HMG1) or ∼20 acidic residues (HMG2). The acidic tail interacts primarily with box B but functions to decrease the DNA-binding affinity of both domains (94, 111, 112). Furthermore, the tail is required for preferential binding to DNA minicircles relative to linear DNA (113). A dynamic assembly in which the acidic tail transiently brings the two HMG box domains together has been proposed. On account of the ability to bend DNA, HMGB proteins are generally referred to as architectural, creating nucleoprotein complexes in which the modified DNA structure promotes the association of additional proteins. In this capacity, HMGB proteins participate in numerous DNA-dependent functions, ranging from DNA replication to gene transcription.
Binding of HMGB proteins to nucleosomes.
Consistent with the preferred binding to four-way junction DNA in vitro in the open-square conformation that is preferred in the absence of divalent cations (114), HMGB proteins bind nucleosomes at DNA entry/exit points (Fig. 2). The DNA bending and underwinding that result from HMGB1 binding are transmitted to the nucleosome core (Fig. 4). This may affect contacts between DNA and core histones and prime the nucleosome core for binding of transcription factors or chromatin-remodeling complexes; thus, HMGB binding is generally associated with more dynamic chromatin and facilitated transcription. HMGB1 binding in the vicinity of the DNA entry/exit points on the nucleosome may be facilitated by interactions between its acidic tail and the N-terminal tail of histone H3, which exits near the DNA entry/exit points of the nucleosome and contacts the linker DNA (Fig. 1) (112, 115–119). Binding of the HMG boxes to DNA frees the acidic tail from intramolecular interactions with the DNA binding surfaces, allowing it to interact with H3. A predicted consequence of this interaction is enhanced DNA binding by HMGB1 (120).
Although mammalian H1 has a high affinity for reconstituted dinucleosomes compared to HMGB1, the binding of HMGB1 displaces the linker histone, perhaps aided by its preferred binding to constrained DNA conformations (121–124). In vitro, an interaction between the acidic tail of HMGB1 and the linker histone H1 has also been reported, suggesting that interactions with H1 may increase the DNA binding affinity of HMGB1 by preventing interactions between the acidic tail and the HMG domains, thereby facilitating the replacement of H1 with HMGB1 (125). This enhanced binding of HMGB1 in turn affects chromatin remodeling, for example, by facilitating the binding of the ISWI-containing remodeling factors ACF and CHRAC to chromatin (115).
YEAST HMGB PROTEIN HMO1
S. cerevisiae expresses several HMGB proteins, of which HMO1 and HMO2 contain two globular HMG-like domains. HMO2 (also known as NHP10), which is unique in exhibiting a preferred binding to DNA ends, is a component of the INO80 chromatin-remodeling complex that is recruited to DNA damage sites (126, 127). HMO1 was first identified by its copurification with an unidentified DNA helicase (128). HMO1 has also been identified in closely related species such as Saccharomyces kluyveri (129). In addition, an HMO1 counterpart is encoded in the Schizosaccharomyces pombe genome (130). According to the Saccharomyces Genome Database (131), HMO1 has been reported to exhibit physical or genetic interactions with a total of 290 genes or gene products. HMO1 is quite abundant, with reported estimates of 19,000 to 25,000 molecules/cell, corresponding to 1 HMO1 molecule per 3 to 4 nucleosomes (58, 60).
HMO1 Domain Organization
HMO1 has two globular domains, named box A and box B, of about 80 amino acids each, similarly to mammalian HMGB (Fig. 1). Box A, which has only limited similarity to consensus HMG domains, functions as a dimerization domain; it has a low affinity for DNA but exhibits some structural specificity, including preferred binding to four-way junction DNA, whereas canonical box B has a higher affinity for DNA but a lower structural specificity (130, 132). The HMO1 box A domain contributes to DNA bending; in contrast, the box B domain contributes most of the DNA binding affinity but fails to bend linear DNA (133). There is no high-resolution structural information available for HMO1; however, a structure-based model predicts that both box A and box B domains adopt the HMG fold (Fig. 5A). Alignment with the human HMGB1 that was used as a template for modeling predicts that Leu55 at the end of helix II corresponds to the HMGB1 box A-intercalating residue (Phe in HMGB1); due to poor sequence conservation at the start of box A, it cannot be predicted with confidence if a potential intercalating residue is present at the end of helix I (Phe at the end of HMGB1 box A helix II is the only intercalating residue in this domain). For HMGB1 box B, Phe and Ile are the DNA-intercalating residues found at the ends of helices I and II, respectively; the corresponding residues in HMO1 box B are Phe114 and Ser138 (which is not predicted to intercalate between DNA bases).
In addition to the A and B domains, HMO1 has a C-terminal domain that is characterized by a stretch of basic amino acids; this is in marked contrast to the mammalian HMGB protein, in which the C-terminal extension is acidic. Deletion of the lysine-rich extension does not reduce the affinity for linear DNA, arguing against a direct interaction between the CTD and this type of DNA substrate. Instead, interactions between box A and the C-terminal extension were reported to induce a conformation that is required for in-phase DNA bending in vitro (133, 134).
Deletion of the HMO1 gene is not lethal but results in a severe growth defect and reduced plasmid stability (128, 135). Inactivation of HMO1 is synthetically lethal with fpr1Δ, a deletion that also results in a plasmid loss phenotype; FPR1 encodes the peptidyl-prolyl cis-trans isomerase FKBP12, and overproduction of HMO1 in cells deleted for FPR1 is toxic. FKBP12 disrupts the self-association of HMO1, suggesting that toxicity could be due to either the uncontrolled accumulation of HMO1 at certain target DNA sites or the sequestering of unbound HMO1 (136). FKBP12 is otherwise best known as the receptor for the immunosuppressive drugs FK506 and rapamycin, and binding to either drug is toxic due to the inhibition of signal transduction (137).
HMO1 Is a Component of the Pol I Transcription Machinery
Transcription of rRNA genes by Pol I has been suggested to be the rate-limiting step in ribosome production (138, 139). Intricate networks adapt rRNA production to metabolic rates, as ribosome production must keep up with cellular demands. The synthesis of rRNA, which accounts for at least 60% of the total transcriptional activity during normal growth, is in most cases thought to be regulated based on the control of active genes as opposed to epigenetic mechanisms that change the ratio of active to silenced genes (140, 141). Distinct nuclear compartments, the nucleoli, form around the rDNA, and nucleolar structure and cell cycle progression are dependent on rDNA transcription (142, 143).
HMO1 associates with rDNA.
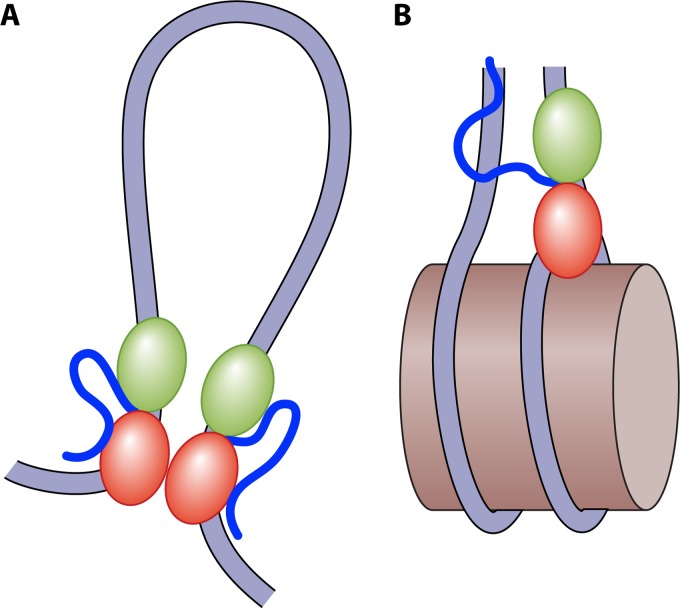
In S. cerevisiae, ∼150 rDNA repeats are arranged head to tail on chromosome XII. Each repeat encodes 35S rRNA synthesized by RNA Pol I and the Pol III-transcribed 5S rRNA. In exponentially growing cells, more than half of the rDNA is transcriptionally silenced (144–147). The remaining fraction constitutes active rRNA genes that are largely depleted of nucleosomes but instead loaded with RNA Pol I and HMO1, with HMO1 stabilizing the open chromatin state in the absence of RNA Pol I transcription (145, 148). While an initial analysis suggested that HMO1 was bound throughout the rDNA (149), a more stringent approach revealed preferred HMO1 binding to Pol I-transcribed regions of the rDNA and that HMO1 remained bound in the absence of Pol I (148, 150). A mechanism by which HMO1 may secure a nucleosome-free region of rDNA involves the dimerization of HMO1 through its box A domains to stabilize DNA bridges and loops (Fig. 6) (151). However, the looped DNA structure formed by HMO1 is dynamic and is predicted to be easily disrupted by the force generated by a transcribing RNA polymerase (151). HMO1 has also been implicated in the resumption of RNA Pol I transcription elongation and reopening of rDNA chromatin after DNA repair; UV light-induced DNA lesions block transcription and lead to a special chromatin structure at the rDNA locus characterized by the dissociation of RNA Pol I and loading of histones downstream of the lesion but retention of HMO1 (152).
FIG 6.
HMO1-mediated stabilization of genomic DNA. (A) On nucleosome-free DNA, HMO1 promotes the formation of loops and bridges that depend on the dimerization of the box A domains (151). Such topological domains may also be mediated by the concerted action of HMO1 and Top2 (195). (B) Both HMO1 and HMO1 deleted for its C-terminal domain can compete with human H1 for binding to nucleosomes, suggesting that HMO1 and H1 binding sites at least partially overlap (198). A possible nucleosome-stabilizing binding mode for HMO1 is illustrated, in which the structure-specific box A domain binds near the dyad and DNA bending by HMO1 is prevented due to the lysine-rich CTD contacting linker DNA.
Upon nutrient limitation, rDNA transcription is downregulated, and this correlates with a reduction in nucleolar size, a process that is dependent on condensins and involves a compaction of the rDNA (153). It was recently reported that HMO1 is also involved in such a contraction of the nucleolus and that its binding is increased across the 35S rRNA gene in response to starvation (154). As noted above, several previous studies have shown HMO1 binding either across the rDNA or with a preferred association with transcribed regions, depending on method of detection, ChIP protocol, and normalization strategy (148–150, 155). In contrast, Wang et al. (154) reported limited HMO1 binding to 35S rRNA genes in log-phase cells (4 h of growth following inoculation of cultures) and an ∼6-fold enrichment during nutrient limitation (24 h of growth); whether the failure to detect HMO1 binding in log-phase cells is due to variations in ChIP protocols or to the genetic background is not clear. The basis for these differences notwithstanding, the reported increase in HMO1 binding upon nutrient limitation would be consistent with a contribution of HMO1 to rDNA compaction under such conditions.
A functional equivalent of UBF.
HMO1 preferentially associates with the transcribed region of the 35S rDNA locus, and it promotes rRNA production both as a component of the RNA Pol I transcription apparatus and by facilitating rRNA maturation (149, 150, 155, 156). Overproduction of HMO1 suppresses the severe growth phenotype caused by a deletion of the gene encoding Rpa49p, a conserved subunit of RNA Pol I and the homolog of human PAF53, and rpa49Δ-hmo1Δ double mutants are inviable, indicating that HMO1 is a component of the Pol I transcription machinery (156). Rrn3p is required for initiation by yeast RNA Pol I, and it is subsequently released during elongation in a process that requires Rpa49p and the presence of another transcribing RNA polymerase (157). The absence of Rpa49p also leads to a decreased density of transcribing RNA polymerases on a given gene, a consequence of which is compromised assembly of the nucleolus (158). The increased distance between transcribing polymerases in the rpa49Δ mutant would also be expected to result in a topological strain due to positive DNA supercoiling accumulating in front of a polymerase and negative supercoiling developing in its wake; consistent with an rpa49Δ mutant accumulating torsional stress, rpa49Δ is lethal when the type I topoisomerase Top3 is inactivated (156). Since HMO1 bends and loops DNA, it may counteract the torsional stress imposed by transcribing Pol I, thereby alleviating the rpa49Δ phenotype. Alternatively, or in addition, the absence of HMO1-mediated DNA looping on rDNA not associated with transcribing RNA polymerase may lead to nucleosome deposition that is inhibitory to transcription (145).
In mammals, RNA Pol I requires upstream binding factor (UBF) for initiation and elongation (159–161). UBF contains six HMG boxes and binds throughout the rRNA gene locus (162). However, yeast lacks UBF, and HMO1 has been proposed to be a functional analog of UBF and to be important for maximal Pol I transcription (156). Comparable functions of UBF and HMO1 are supported by the observation that both proteins are highly enriched in the nucleolus and localized throughout the transcribed rDNA region and that both proteins contain HMG domains that may promote DNA bending and DNA looping (149, 150, 162, 163). Further support for overlapping functions of HMO1 and UBF was provided by the observation that the expression of human UBF1 or S. pombe HMO1 also suppresses the rpa49Δ growth phenotype (130).
HMO1 Regulates Pol II Transcription
Ribosomal protein gene expression.
In S. cerevisiae, the ribosome is made up of four rRNAs (5S, 5.8S, 18S, and 25S) and 79 ribosomal proteins (RPs) expressed from 138 genes (164). RP gene transcription constitutes up to 50% of RNA Pol II-mediated transcription, and it is coordinately regulated in response to environmental conditions (165). In prokaryotes, ensuring the production of stoichiometric levels of ribosomal proteins is simple because RP genes form operons, whereas in eukaryotes, such regulation is more complicated, as each RP gene yields a monocistronic mRNA. A number of transcription factors have been reported to contribute to the regulation of RP gene activity, including Rap1p, which binds the majority of RP genes and forms nucleosome-free regions in target promoters (155, 166). HMO1 binds RP gene promoters with variable occupancy and has been implicated in preinitiation complex (PIC) assembly by covering a nucleosome-free region and in the recruitment of the transcription factor Fhl1p (forkhead like) (149, 150, 155, 167, 168).
Pol II transcription requires basal transcription factors, including TFIID, which contains the TATA box binding protein (TBP), and TBP-associated factors (TAFs). The TAF1 N-terminal domain (TAND) inhibits the binding of TBP to the TATA element (169); it has been reported that HMO1 interacts with TBP and TAND and that HMO1 deletion decreases the transcription of TAND-dependent genes, suggesting that HMO1 prevents inhibitory TBP-TAND interactions. In addition, an interaction between HMO1 and TFIID was suggested by the observation that HMO1 deletion causes an upstream shift in transcription start sites of genes under the control of HMO1-enriched promoters but not of genes under the control of promoters with limited HMO1 occupancy (168). This shift in the transcriptional start site was subsequently linked to the ability of HMO1 to mask a nucleosome-free region to prevent inappropriate PIC assembly (167). This nucleosome-free region was later reported to exhibit sensitivity to micrococcal nuclease and to contain unstable or “fragile” nucleosomes, perhaps rendered unstable through the action of the essential multifunctional transcription factor Rap1p (170).
Fhl1p is a transcription factor with sequence similarity to the forkhead (FH) winged-helix DNA binding domain. On RP genes, Fhl1p has been reported to remain bound and to recruit either the coactivator Ifh1p (interacts with forkhead) or the corepressor Crf1p. During vigorous growth, Fhl1p and Rap1p recruit Ifh1p, which results in maximal transcription (171–173). In addition, Sfp1p (split finger protein) has been reported to be required for maximal transcription from RP promoters (174). During stress and nutrient starvation, Ifh1p dissociates from RP promoters, and Fhl1p recruits Crf1p, while Sfp1p translocates to the cytoplasm, events that lead to the downregulation of RP gene transcription (171, 172, 174, 175). Dissociation of HMO1 was also reported under conditions of RP gene repression, leading to an upstream shift of the +1 nucleosome, suggesting that HMO1 is important for the placement of the +1 nucleosomes in either a repressive or an active position (176).
Regulation of non-RP gene transcription.
Little is known about the role of HMO1 in the regulation of Pol II-transcribed genes other than the RP genes. Excess HMO1 represses the HMO1 promoter; however, the underlying mechanism is unknown (135). Given the self-association of HMO1, it is tempting to speculate that excess HMO1 promotes an accretion of HMO1 on the HMO1 promoter that adversely affects the binding of either transcription factors or RNA Pol II. In contrast, the absence of HMO1 results in the reduced transcription of the MATa gene, a locus to which HMO1 also binds directly, suggesting that the effect may be direct (177).
Regulation of gene transcription involves ATP-dependent chromatin-remodeling complexes that either slide or evict nucleosomes or alter their composition (178). The conserved SWI/SNF complex, for instance, is critical for the modulation of gene expression during a variety of cellular processes. Among the HMGB proteins, HMO1 and NHP6A/B stimulate the sliding activity of SWI/SNF, but only HMO1 promotes SWI/SNF binding to the nucleosome, histone octamer transfer, and exposure of nucleosomal DNA. Notably, the stimulatory effect requires the HMO1 CTD and the presence of linker DNA, as no binding of HMO1 to nucleosomes devoid of linker DNA could be detected (179). Based on these observations, HMO1 appears to recruit SWI/SNF to nucleosomes by a mechanism that requires changes in the DNA topology or organization of the nucleosomal array.
HMO1 Coordinates Responses to TORC1 Signaling
Control of rRNA synthesis.
Cell growth requires ribosome biogenesis, and several signaling pathways participate by sensing environmental cues such as stress, nutrient limitation, or growth factors. The serine/threonine protein kinase TOR (target of rapamycin) is a central regulator of ribosome biogenesis. Both yeast and mammalian cells have two functionally different TOR complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2); TORC1 is stimulated by amino acid sufficiency, and it responds to rapamycin to generate responses akin to those elicited by starvation and environmental stress such as hypoxia and DNA damage to reduce cell growth (180, 181). TORC2 is rapamycin insensitive and controls the organization of the actin cytoskeleton (181–183).
In addition to targeting the translation machinery, TORC1 promotes the transcription of genes whose products are involved in ribosome biogenesis. TOR kinase is recruited directly to rRNA gene promoters in a nutrient-dependent fashion; in the presence of rapamycin, rDNA becomes condensed (and the nucleolus contracts by a mechanism in which HMO1 participates, as noted above), and a rapid delocalization of RNA Pol I from the nucleolus is observed, along with TOR kinase exiting the nucleus (184, 185). Rapamycin treatment has also been reported to result in the dissociation of HMO1 from rDNA, and TORC1-dependent repression of rDNA transcription is reduced in the absence of HMO1, suggesting a role for HMO1 in communicating TORC1 activity (150). Nutrient limitation is a condition expected to inhibit TORC1; as noted above, a recent study suggested that nutrient limitation (achieved by growing yeast cells for 24 h following inoculation) resulted in increased HMO1 binding to rDNA and to nucleolar contraction (154). It would be instructive to determine TORC1 activity after 24 h of growth compared to the more extensive depletion of nutrients and to investigate if increased HMO1 binding to rDNA may be a transient phenomenon. Prolonged exposure to rapamycin results in the downregulation of the HMO1 promoter and reduced cellular HMO1 levels (135, 186), perhaps rationalizing an initial increase in the HMO1 association with rDNA upon the inhibition of TORC1 signaling, followed by its dissociation as cellular levels decrease.
TORC1 signaling has also been reported to control histone H3 lysine 56 acetylation (H3K56ac), with H3K56ac promoting the binding of HMO1 (187). H3K56ac is generally associated with the disassembly of nucleosomes by disrupting interactions between H3 and DNA where DNA enters and exits the nucleosome (188). Accordingly, H3K56ac may promote a nucleosome-free region in rDNA to which HMO1 is recruited, while dissociation of HMO1 from rDNA upon the inhibition of TORC1 may be linked to reduced H3K56ac.
Control of RP gene expression.
TORC1 also controls RP gene expression. For example, inhibition of TORC1 signaling by rapamycin releases the histone acetyltransferase ESA1 from RP gene promoters and leads to histone H4 deacetylation (184). Several transcription factors have been implicated in the regulation of RP gene activity. At least part of the rapamycin-mediated downregulation of RP gene activity can be attributed to the accumulation of Crf1p in the nucleus, an event that is triggered by its phosphorylation in response to nutrient limitation (171, 173, 189, 190). TORC1 signaling also leads to the accumulation of Sfp1p in the nucleus (174, 175). Under optimal growth conditions, Sfp1p binds RP gene promoters and activates transcription, perhaps via an interaction with Fhl1p and Ifh1p, while inhibition of TORC1 signaling results in the release of Sfp1p and its exit from the nucleus (171, 174, 175). HMO1 has also been reported to dissociate from some RP gene promoters in response to rapamycin, and reduced TORC1-dependent repression of RP gene expression was observed in hmo1Δ cells (150). Notably, the expression of HMO1 is also repressed upon the inhibition of TORC1 signaling in an HMO1- and Fhl1p-dependent fashion, suggesting a mechanism comparable to that proposed for the TORC1-mediated repression of RP genes (135). Clearly, HMO1 is linked to the TORC1 signaling pathway and may be involved in coordinating rDNA transcription and RP gene expression (150, 191).
HMO1 Stabilizes Noncanonical Chromatin Structures
HMO1 stabilizes repeat tracts.
On rDNA and on ribosomal protein gene promoters, HMO1 appears to exert an effect in large part through its association with nucleosome-free DNA or DNA associated with fragile nucleosomes. The potential instability of DNA containing repetitive sequence elements, such as that characterizing the rDNA array, necessitates protective measures. In humans, long CAG repeat tracts underlie hereditary neurodegenerative diseases, including Huntington disease, as they have a propensity to expand. The length of CAG repeat tracts correlates with their instability; duplex DNA exhibits unusual flexibility, and unwound DNA may engage in intramolecular base pairing to form hairpin structures that hinder DNA replication (192). When embedded in the yeast chromosome, CAG repeat tracts are bound and stabilized by HMO1, which establishes a noncanonical chromatin organization (193). The length of CAG repeat tract chromatin that is protected from nuclease digestion is shorter than that protected by a nucleosome, raising the possibility that tetramer cores of histones associate with the DNA and that HMO1 may have replaced H2A and H2B, perhaps serving as a linker between tetramer cores.
Replication fork encounters with transcription.
Recombination events and genomic instability may also be triggered by clashes between replication and transcription (194). Dedicated topoisomerases such as Top2p relieve the topological constraints that result when a replication fork encounters transcription and promote fork progression. In S phase, intergenic regions close to some transcribed genes exhibit low nucleosome density but accumulate both HMO1 and Top2p; together, Top2p and HMO1 appear to suppress chromosome fragility at the M/G1 transition (195). Top2p binding occurs independently of HMO1, while a function of HMO1 may be to maintain the low nucleosome density required to facilitate Top2p-mediated DNA looping, which promotes the formation of topological domains and gene transcription.
DNA damage response.
DNA damage may lead to events ranging from mutagenesis to chromosomal rearrangements. The DNA damage response (DDR) allows for a delay of cell cycle progression to ensure DNA repair and replication of the genome by high-fidelity polymerases and to mediate fork restart (196). The error-free mode involves a recombination event in which the newly synthesized strand is used as the template for replication of the damaged strand, whereas the error-prone mode relies on trans-lesion synthesis. This pathway choice is important for genome integrity. Among myriad events associated with replication, DNA topological changes include the sister chromatid bridges that form when replication forks pass through transcriptionally active chromatin loops (195). The association of HMO1 with such junctions has also been implicated as one of the mechanisms by which HMO1 promotes the error-free DNA damage tolerance pathway by facilitating template switching (197), and it is consistent with the preferred binding of HMO1 to four-way DNA junctions compared to linear DNA (132). These functions of HMO1 require its C-terminal extension (197), shown to be required for DNA bending and bridging (133, 134).
HMO1 and DNA Double-Strand Break Repair
HMO1 stabilizes chromatin.
The absence of HMO1 does not affect the bead-and-string pattern of nucleosomes but makes the chromatin hypersensitive to nuclease (128, 198), suggesting that HMO1 stabilizes chromatin. A genome-wide analysis of HMO1 binding revealed extensive yet variable associations across the genome, with particular enrichment at genes encoding ribosomal proteins and rRNA (149). The coverage of HMO1 binding would be consistent with an effect on chromatin stabilization that is detectable by analysis of bulk chromatin.
DSBs are induced in the context of chromatin. The presence of sister chromatids allows repair by homologous recombination, whereas nonhomologous end joining operates without involving a separate copy of the DNA duplex. A number of histone modification and chromatin-remodeling events precede DNA repair pathway choice and render the chromatin template accessible to repair proteins (199). In yeast, a DSB may be site-specifically created at the mating-type locus MAT by inducing the expression of HO endonuclease. HMO1 binds this locus, and events involved in DSB repair include the eviction of HMO1 proximal to the DSB site (177). Notably, events such as histone H2A phosphorylation, INO80 recruitment, and nucleosome disruption (as evidenced by H3 eviction) occur faster in the absence of HMO1, suggesting that the absence of HMO1 creates a more accessible chromatin state. The facilitated chromatin remodeling is associated with more efficient DNA end resection, recruitment of DNA repair proteins, and repair by both homologous recombination and nonhomologous end joining. Consistent with the roles of HMO1 in maintaining nucleosome-free DNA segments, restoration of the original chromatin state also occurs faster in the absence of HMO1 (177).
Replication-independent endogenous DNA DSBs occur spontaneously and are not pathological lesions in that they do not induce mutation or cell death. Instead, they may possess important biological functions, perhaps in relieving topological stress that might otherwise result in uncontrolled DNA breakage. They have also been reported to occur nonrandomly, for example, with an increased frequency in heterochromatin (200). Such breaks are repaired by either Ku- or Rad51p-dependent pathways, as evidenced by the increased levels of such breaks in cells deleted for either Ku or Rad51p (201). Conversely, deletion of HMO1 resulted in reduced break levels, an outcome that would be consistent with the absence of HMO1 facilitating chromatin-remodeling events required for their elimination.
The HMO1 C-terminal domain is required for chromatin stabilization.
The finding that chromatin-remodeling events associated with DSB repair occur faster in the absence of HMO1 points to a role for HMO1 in stabilizing chromatin. Notably, this stabilization requires the lysine-rich C-terminal domain, which was previously shown to be required for bridging separate DNA strands (134, 177). A role for the basic extension in chromatin stabilization is corroborated by the increased sensitivity to MNase characteristic of chromatin isolated from both hmo1Δ cells and cells expressing HMO1 deleted for its C-terminal extension (198). Notably, the expression of human histone H1 in hmo1Δ cells restores wild-type levels of MNase sensitivity, and it also complements the HMO1 deletion in terms of the faster chromatin-remodeling events characteristic of hmo1Δ cells (198). HMO1 deleted for its lysine-rich CTD still binds chromatin and can compete with H1 for binding. Taken together, these observations suggest that HMO1 can fulfill the role of a linker histone and that it shares with mammalian H1 the requirement for a lysine-rich domain for chromatin stabilization to be manifest.
While the exact nature of the HMO1 interaction with nucleosomes remains unknown, the preferred binding of HMO1 to four-way DNA junctions is consistent with binding at the nucleosome dyad, as reported for H1, which likewise binds preferentially to DNA junctions (Fig. 2) (38). Based on the inference that DNA bending and underwinding by mammalian HMGB may facilitate nucleosome unwrapping, HMO1 might likewise be expected to destabilize nucleosomes; however, the observation that interactions between the HMO1 box A domain and the CTD are required for DNA bending offers an alternative scenario (118, 133, 134). It is conceivable that the HMO1 lysine-rich domain contacts DNA directly when HMO1 associates with nucleosomes, a circumstance in which DNA bending by HMO1 would likely be attenuated (Fig. 6), thus allowing HMO1 to stabilize the nucleosome. Thus, we propose a model in which HMO1 binds near the nucleosome dyad axis, most likely with its box A domain, which has the greatest preference for four-way junction DNA; such an interaction might free the lysine-rich CTD to interact with linker DNA, preventing DNA bending and generating a compacted nucleosome array with limited MNase access.
Interplay between Hho1p and HMO1
Both genes encoding core histones and the HHO1 gene are transcribed in S phase, suggesting that Hho1p acts in concert with the core histones (202). Consistent with this observation, Hho1p binds the DNA entry/exit points of nucleosomes (56). However, during vegetative growth, the absence of Hho1p does not affect global chromatin structure, as evidenced by changes in MNase sensitivity (56, 198). A contributing factor may be its relatively low abundance, estimated to be 1 Hho1p molecule per 37 nucleosomes, a cellular content that is ∼10-fold lower than that of HMO1 (57, 58, 60). Furthermore, inactivation of HHO1 does not result in growth or mating defects, significant global changes in gene expression, or a change in average nucleosome distance (57). Phenotypes associated with HHO1 deletion are subtle and have pointed to roles for Hho1p in suppressing homologous recombination and the formation of silent chromatin (perhaps by affecting the function of the Sir complex), in promoting the formation of chromatin loops, and in chromatin compaction during stationary phase (59, 62–64). Disruption of HHO1 results in a substantial increase in the levels of its own transcript, suggesting a feedback system for HHO1 gene regulation (57). Curiously, a feedback mechanism for the regulation of the HMO1 promoter was also suggested by the increased HMO1 promoter activity in an HMO1Δ strain (135).
Another commonality between Hho1p and HMO1 is their preferred binding to rDNA. For Hho1p, this localization to rDNA is associated with a repression of recombination and with efficient transcriptional silencing by the compaction of rDNA chromatin (61, 85), whereas functions of HMO1 range from rDNA compaction during starvation to participating as a component of the Pol I transcription machinery, as discussed above (149, 150, 152, 154–156). While the inactivation of HHO1 causes a modest enrichment of HMO1 at rDNA, the absence of HMO1 results in a significant increase in the association of Hho1p (198). The finding that the absence of either HMO1 or Hho1p results in a reciprocal increase in the binding of the other protein at rDNA may explain why the elimination of both proteins is required to render rDNA significantly more susceptible to MNase digestion. This interdependence appears to be specific to rDNA; for example, both HMO1 and Hho1p bind the mating-type locus MAT, yet the absence of one protein does not affect the binding of the other, and only HMO1 can protect linker DNA from MNase digestion at this locus (198).
CONCLUSIONS
The evolutionarily variable linker histones are critical for the stabilization of nucleosomes by binding DNA entering and exiting the core particle and by facilitating higher-order organization (15). The lysine-rich CTD of metazoan H1 is crucial for such stabilization; indeed, single-domain linker histones corresponding to the CTD are, for example, found in kinetoplastids, and even eubacteria have been shown to compact their genomic DNA by means of proteins with low-complexity lysine-rich extensions (45, 203). The lysine-rich C-terminal domain of HMO1 is unique for HMGB proteins and in marked contrast to mammalian HMGB proteins (and even yeast HMO2) that have acidic tails. The absence of HMO1 or deletion of the HMO1 CTD makes chromatin hypersensitive to nuclease, and it facilitates chromatin-remodeling events associated with DSB repair phenotypes that are complemented by the expression of human H1 in the hmo1Δ strain, pointing to a role for HMO1 in chromatin stabilization. Establishing the nature of the HMO1 interaction with reconstituted nucleosomes will be important to shed further light on the mechanism by which HMO1 executes such H1-like nucleosome stabilization. However, the ability of HMO1 to stabilize genomic DNA appears to go beyond conventional linker histone function. HMO1 plays a role in transcription by both RNA Pol I and Pol II, and it functions in the DNA damage response by directing lesions toward the error-free pathway. In these circumstances, HMO1 is required for the stabilization of nucleosome-free DNA or DNA associated with fragile nucleosomes. Taken together, emerging data suggest that yeast linker histone function is a division of labor between Hho1p and HMO1 in which Hho1p may have acquired more specialized functions due to its unusual domain organization, whereas the terminal lysine-rich extension of HMO1 has endowed it with the ability to stabilize both noncanonical and conventional nucleosome arrays.
ACKNOWLEDGMENTS
We are grateful for awards from the National Science Foundation (MCB-1051610 and MCB-1515349) and an LSU faculty research grant (to A.G.).
REFERENCES
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Schalch T, Duda S, Sargent DF, Richmond TJ. 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 3.Thastrom A, Bingham LM, Widom J. 2004. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol 338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Wu B, Mohideen K, Vasudevan D, Davey CA. 2010. Structural insight into the sequence dependence of nucleosome positioning. Structure 18:528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Smith S, Stillman B. 1991. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J 10:971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatakeyama A, Hartmann B, Travers A, Nogues C, Buckle M. 2016. High-resolution biophysical analysis of the dynamics of nucleosome formation. Sci Rep 6:27337. doi: 10.1038/srep27337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. 2009. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol 16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Koning L, Corpet A, Haber JE, Almouzni G. 2007. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, D'Arcy S, Radebaugh CA, Krzizike DD, Giebler HA, Huang L, Nyborg JK, Luger K, Stargell LA. 2016. Histone chaperone Nap1 is a major regulator of histone H2A-H2B dynamics at the inducible GAL locus. Mol Cell Biol 36:1287–1296. doi: 10.1128/MCB.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguilar-Gurrieri C, Larabi A, Vinayachandran V, Patel NA, Yen K, Reja R, Ebong IO, Schoehn G, Robinson CV, Pugh BF, Panne D. 2016. Structural evidence for Nap1-dependent H2A-H2B deposition and nucleosome assembly. EMBO J 35:1465–1482. doi: 10.15252/embj.201694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. 2013. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface 10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghirlando R, Felsenfeld G. 2008. Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J Mol Biol 376:1417–1425. doi: 10.1016/j.jmb.2007.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu R-M, Zhu P, Li G. 2014. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 14.Hergeth SP, Schneider R. 15 October 2015. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harshman SW, Young NL, Parthun MR, Freitas MA. 2013. H1 histones: current perspectives and challenges. Nucleic Acids Res 41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan TW, Brown DT. 2016. Molecular dynamics of histone H1. Biochim Biophys Acta 1859:468–475. doi: 10.1016/j.bbagrm.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Routh A, Sandin S, Rhodes D. 2008. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A 105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T. 2012. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J 31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160:1145–1158. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh T-HS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. 2015. Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigoryev SA, Bascom G, Buckwalter JM, Schubert MB, Woodcock CL, Schlick T. 2016. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc Natl Acad Sci U S A 113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai R, Komeda S, Shimura M, Tamura S, Matsuyama S, Nishimura K, Rogge R, Matsunaga A, Hiratani I, Takata H, Uemura M, Iida Y, Yoshikawa Y, Hansen JC, Yamauchi K, Kanemaki MT, Maeshima K. 2016. Chromatin folding and DNA replication inhibition mediated by a highly antitumor-active tetrazolato-bridged dinuclear platinum(II) complex. Sci Rep 6:24712. doi: 10.1038/srep24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S. 2010. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci U S A 107:9620–9625. doi: 10.1073/pnas.1000309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivolob A, Prunell A. 2003. Linker histone-dependent organization and dynamics of nucleosome entry/exit DNAs. J Mol Biol 331:1025–1040. doi: 10.1016/S0022-2836(03)00831-3. [DOI] [PubMed] [Google Scholar]
- 25.An W, van Holde K, Zlatanova J. 1998. The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. J Biol Chem 273:26289–26291. doi: 10.1074/jbc.273.41.26289. [DOI] [PubMed] [Google Scholar]
- 26.Bednar J, Hamiche A, Dimitrov S. 2016. H1-nucleosome interactions and their functional implications. Biochim Biophys Acta 1859:436–443. doi: 10.1016/j.bbagrm.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 28.Lever MA, Th'ng JP, Sun X, Hendzel MJ. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 29.Woodcock CL, Skoultchi AI, Fan Y. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 30.Ponte I, Vila R, Suau P. 2003. Sequence complexity of histone H1 subtypes. Mol Biol Evol 20:371–380. doi: 10.1093/molbev/msg041. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan V, Finch J, Graziano V, Lee P, Sweet R. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhou BR, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y. 2013. Structural insights into the histone H1-nucleosome complex. Proc Natl Acad Sci U S A 110:19390–19395. doi: 10.1073/pnas.1314905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou BR, Jiang J, Feng H, Ghirlando R, Xiao TS, Bai Y. 2015. Structural mechanisms of nucleosome recognition by linker histones. Mol Cell 59:628–638. doi: 10.1016/j.molcel.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George EM, Izard T, Anderson SD, Brown DT. 2010. Nucleosome interaction surface of linker histone H1c is distinct from that of H1(0). J Biol Chem 285:20891–20896. doi: 10.1074/jbc.M110.108639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown DT, Izard T, Misteli T. 2006. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varga-Weisz P, van Holde K, Zlatanova J. 1993. Preferential binding of histone H1 to four-way helical junction DNA. J Biol Chem 268:20699–20700. [PubMed] [Google Scholar]
- 37.Hendzel MJ, Lever MA, Crawford E, Th'ng JP. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 38.White AE, Hieb AR, Luger K. 2016. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci Rep 6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang H, Clark DJ, Hayes JJ. 2012. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res 40:1475–1484. doi: 10.1093/nar/gkr866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark D, Hill C, Martin S, Thomas J. 1988. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J 7:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caterino TL, Hayes JJ. 2011. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol 89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque A, Collepardo-Guevara R, Grigoryev S, Schlick T. 2014. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res 42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. 1986. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol 187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 44.Vyas P, Brown DT. 2012. N-and C-terminal domains determine differential nucleosomal binding geometry and affinity of linker histone isotypes H10 and H1c. J Biol Chem 287:11778–11787. doi: 10.1074/jbc.M111.312819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasinsky HE, Lewis JD, Dacks JB, Ausio J. 2001. Origin of H1 linker histones. FASEB J 15:34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 46.Millan-Arino L, Izquierdo-Bouldstridge A, Jordan A. 2016. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta 1859:510–519. doi: 10.1016/j.bbagrm.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 47.Izzo A, Schneider R. 2016. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta 1859:486–495. doi: 10.1016/j.bbagrm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Galanti N, Galindo M, Sabaj V, Espinoza I, Toro G. 1998. Histone genes in trypanosomatids. Parasitol Today 14:64–70. doi: 10.1016/S0169-4758(97)01162-9. [DOI] [PubMed] [Google Scholar]
- 49.Doenecke D, Tönjes R. 1986. Differential distribution of lysine and arginine residues in the closely related histones H1 and H5: analysis of a human H1 gene. J Mol Biol 187:461–464. doi: 10.1016/0022-2836(86)90446-8. [DOI] [PubMed] [Google Scholar]
- 50.Ushinsky S, Bussey H, Ahmed A, Wang Y, Friesen J, Williams B, Storms R. 1997. Histone H1 in Saccharomyces cerevisiae. Yeast 13:151–161. [DOI] [PubMed] [Google Scholar]
- 51.Ono K, Kusano O, Shimotakahara S, Shimizu M, Yamazaki T, Shindo H. 2003. The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy. Nucleic Acids Res 31:7199–7207. doi: 10.1093/nar/gkg931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali T, Coles P, Stevens TJ, Stott K, Thomas JO. 2004. Two homologous domains of similar structure but different stability in the yeast linker histone, Hho1p. J Mol Biol 338:139–148. doi: 10.1016/j.jmb.2004.02.046. [DOI] [PubMed] [Google Scholar]
- 53.Ali T, Thomas JO. 2004. Distinct properties of the two putative “globular domains” of the yeast linker histone, Hho1p. J Mol Biol 337:1123–1135. doi: 10.1016/j.jmb.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 54.Schafer G, Smith EM, Patterton HG. 2005. The Saccharomyces cerevisiae linker histone Hho1p, with two globular domains, can simultaneously bind to two four-way junction DNA molecules. Biochemistry 44:16766–16775. doi: 10.1021/bi0511787. [DOI] [PubMed] [Google Scholar]
- 55.Noll M, Kornberg RD. 1977. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol 109:393–404. doi: 10.1016/S0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 56.Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J Biol Chem 273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 57.Freidkin I, Katcoff DJ. 2001. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res 29:4043–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 59.Downs JA, Kosmidou E, Morgan A, Jackson SP. 2003. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell 11:1685–1692. doi: 10.1016/S1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 60.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 61.Levy A, Eyal M, Hershkovits G, Salmon-Divon M, Klutstein M, Katcoff DJ. 2008. Yeast linker histone Hho1p is required for efficient RNA polymerase I processivity and transcriptional silencing at the ribosomal DNA. Proc Natl Acad Sci U S A 105:11703–11708. doi: 10.1073/pnas.0709403105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schafer G, McEvoy CR, Patterton HG. 2008. The Saccharomyces cerevisiae linker histone Hho1p is essential for chromatin compaction in stationary phase and is displaced by transcription. Proc Natl Acad Sci U S A 105:14838–14843. doi: 10.1073/pnas.0806337105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Georgieva M, Roguev A, Balashev K, Zlatanova J, Miloshev G. 2012. Hho1p, the linker histone of Saccharomyces cerevisiae, is important for the proper chromatin organization in vivo. Biochim Biophys Acta 1819:366–374. doi: 10.1016/j.bbagrm.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Yu Q, Kuzmiak H, Zou Y, Olsen L, Defossez PA, Bi X. 2009. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J Biol Chem 284:740–750. doi: 10.1074/jbc.M806274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Veron M, Zou Y, Yu Q, Bi X, Selmi A, Gilson E, Defossez PA. 2006. Histone H1 of Saccharomyces cerevisiae inhibits transcriptional silencing. Genetics 173:579–587. doi: 10.1534/genetics.105.050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang C, Pugh BF. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, Hicks J, Bryant GO, Ptashne M. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izzo A, Kamieniarz-Gdula K, Ramírez F, Noureen N, Kind J, Manke T, van Steensel B, Schneider R. 2013. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep 3:2142–2154. doi: 10.1016/j.celrep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 69.Bouvet P, Dimitrov S, Wolffe AP. 1994. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev 8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 70.Mayor R, Izquierdo-Bouldstridge A, Millán-Ariño L, Bustillos A, Sampaio C, Luque N, Jordan A. 2015. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J Biol Chem 290:7474–7491. doi: 10.1074/jbc.M114.617324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. 2005. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 72.Parseghian MH, Newcomb RL, Winokur ST, Hamkalo BA. 2000. The distribution of somatic H1 subtypes is non-random on active vs. inactive chromatin: distribution in human fetal fibroblasts. Chromosome Res 8:405–424. doi: 10.1023/A:1009262819961. [DOI] [PubMed] [Google Scholar]
- 73.Kim JM, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W. 2015. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci Rep 5:16714. doi: 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. 2005. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem 280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 75.Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. 2014. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin 7:16. doi: 10.1186/1756-8935-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernier M, Luo Y, Nwokelo KC, Goodwin M, Dreher SJ, Zhang P, Parthun MR, Fondufe-Mittendorf Y, Ottesen JJ, Poirier MG. 2015. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat Commun 6:10152. doi: 10.1038/ncomms10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao K, Lailler N, Zhang Y, Kumar A, Uppal K, Liu Z, Lee EK, Wu H, Medrzycki M, Pan C. 2013. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet 9:e1003417. doi: 10.1371/journal.pgen.1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Q, Giebler HA, Isaacson MK, Nyborg JK. 2015. Eviction of linker histone H1 by NAP-family histone chaperones enhances activated transcription. Epigenetics Chromatin 8:1. doi: 10.1186/1756-8935-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soria G, Polo SE, Almouzni G. 2012. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 80.Smerdon MJ. 1991. DNA repair and the role of chromatin structure. Curr Opin Cell Biol 3:422–428. doi: 10.1016/0955-0674(91)90069-B. [DOI] [PubMed] [Google Scholar]
- 81.Dantuma NP, van Attikum H. 2016. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J 35:6–23. doi: 10.15252/embj.201592595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 83.Alexiadis V, Kadonaga JT. 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev 16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J Biol Chem 278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 85.Li C, Mueller JE, Elfline M, Bryk M. 2008. Linker histone H1 represses recombination at the ribosomal DNA locus in the budding yeast Saccharomyces cerevisiae. Mol Microbiol 67:906–919. doi: 10.1111/j.1365-2958.2007.06101.x. [DOI] [PubMed] [Google Scholar]
- 86.Machida S, Takaku M, Ikura M, Sun J, Suzuki H, Kobayashi W, Kinomura A, Osakabe A, Tachiwana H, Horikoshi Y. 2014. Nap1 stimulates homologous recombination by RAD51 and RAD54 in higher-ordered chromatin containing histone H1. Sci Rep 4:4863. doi: 10.1038/srep04863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roberts SA, Ramsden DA. 2007. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J Biol Chem 282:10605–10613. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- 88.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S. 2015. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 89.Bianchi ME, Agresti A. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 90.Duguet M, de Recondo AM. 1978. A deoxyribonucleic acid unwinding protein isolated from regenerating rat liver. Physical and functional properties. J Biol Chem 253:1660–1666. [PubMed] [Google Scholar]
- 91.Magna M, Pisetsky DS. 2014. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol Med 20:138–146. doi: 10.2119/molmed.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 93.Bianchi ME, Beltrame M, Paonessa G. 1989. Specific recognition of cruciform DNA by nuclear protein HMG1. Science 243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 94.Jung Y, Lippard SJ. 2003. Nature of full-length HMGB1 binding to cisplatin-modified DNA. Biochemistry 42:2664–2671. doi: 10.1021/bi026972w. [DOI] [PubMed] [Google Scholar]
- 95.Masse JE, Wong B, Yen Y-M, Allain FH-T, Johnson RC, Feigon J. 2002. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol 323:263–284. doi: 10.1016/S0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 96.Churchill ME, Klass J, Zoetewey DL. 2010. Structural analysis of HMGD-DNA complexes reveals influence of intercalation on sequence selectivity and DNA bending. J Mol Biol 403:88–102. doi: 10.1016/j.jmb.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gerlitz G, Hock R, Ueda T, Bustin M. 2009. The dynamics of HMG protein-chromatin interactions in living cells. Biochem Cell Biol 87:127–137. doi: 10.1139/O08-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Webb M, Payet D, Lee K-B, Travers AA, Thomas JO. 2001. Structural requirements for cooperative binding of HMG1 to DNA minicircles. J Mol Biol 309:79–88. doi: 10.1006/jmbi.2001.4667. [DOI] [PubMed] [Google Scholar]
- 99.Lange SS, Vasquez KM. 2009. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog 48:571–580. doi: 10.1002/mc.20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grove A, Galeone A, Mayol L, Geiduschek EP. 1996. Localized DNA flexibility contributes to target site selection by DNA-bending proteins. J Mol Biol 260:120–125. doi: 10.1006/jmbi.1996.0386. [DOI] [PubMed] [Google Scholar]
- 101.Huang J-C, Zamble DB, Reardon JT, Lippard SJ, Sancar A. 1994. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci U S A 91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S-I, Asagoshi K, Hou EW, Khodyreva SN. 2007. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell 27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He Q, Liang CH, Lippard SJ. 2000. Steroid hormones induce HMG1 overexpression and sensitize breast cancer cells to cisplatin and carboplatin. Proc Natl Acad Sci U S A 97:5768–5772. doi: 10.1073/pnas.100108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park S, Lippard SJ. 2011. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 50:2567–2574. doi: 10.1021/bi2000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mukherjee A, Vasquez KM. 2016. HMGB1 interacts with XPA to facilitate the processing of DNA interstrand crosslinks in human cells. Nucleic Acids Res 44:1151–1160. doi: 10.1093/nar/gkv1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teo SH, Grasser KD, Thomas JO. 1995. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur J Biochem 230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 107.Assenberg R, Webb M, Connolly E, Stott K, Watson M, Hobbs J, Thomas JO. 2008. A critical role in structure-specific DNA binding for the acetylatable lysine residues in HMGB1. Biochem J 411:553–561. doi: 10.1042/BJ20071613. [DOI] [PubMed] [Google Scholar]
- 108.Paull TT, Haykinson MJ, Johnson RC. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev 7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 109.Stott K, Tang GS, Lee K-B, Thomas JO. 2006. Structure of a complex of tandem HMG boxes and DNA. J Mol Biol 360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 110.Sánchez-Giraldo R, Acosta-Reyes FJ, Malarkey CS, Saperas N, Churchill ME, Campos J. 2015. Two high-mobility group box domains act together to underwind and kink DNA. Acta Crystallogr D Biol Crystallogr 71:1423–1432. doi: 10.1107/S1399004715007452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Knapp S, Müller S, Digilio G, Bonaldi T, Bianchi ME, Musco G. 2004. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry 43:11992–11997. doi: 10.1021/bi049364k. [DOI] [PubMed] [Google Scholar]
- 112.Stott K, Watson M, Howe FS, Grossmann JG, Thomas JO. 2010. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol 403:706–722. doi: 10.1016/j.jmb.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 113.Lee K-B, Thomas JO. 2000. The effect of the acidic tail on the DNA-binding properties of the HMG1, 2 class of proteins: insights from tail switching and tail removal. J Mol Biol 304:135–149. doi: 10.1006/jmbi.2000.4206. [DOI] [PubMed] [Google Scholar]
- 114.Pöhler JR, Norman DG, Bramham J, Bianchi ME, Lilley DM. 1998. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J 17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bonaldi T, Längst G, Strohner R, Becker PB, Bianchi ME. 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J 21:6865–6873. doi: 10.1093/emboj/cdf692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Travers AA. 2003. Priming the nucleosome: a role for HMGB proteins? EMBO Rep 4:131–136. doi: 10.1038/sj.embor.embor741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. 2004. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry 43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
- 118.Joshi SR, Sarpong YC, Peterson RC, Scovell WM. 2012. Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding. Nucleic Acids Res 40:10161–10171. doi: 10.1093/nar/gks815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Watson M, Stott K, Fischl H, Cato L, Thomas JO. 2014. Characterization of the interaction between HMGB1 and H3—a possible means of positioning HMGB1 in chromatin. Nucleic Acids Res 42:848–859. doi: 10.1093/nar/gkt950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol 24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ura K, Nightingale K, Wolffe AP. 1996. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J 15:4959–4969. [PMC free article] [PubMed] [Google Scholar]
- 122.Polanska E, Pospisilova S, Stros M. 2014. Binding of histone H1 to DNA is differentially modulated by redox state of HMGB1. PLoS One 9:e89070. doi: 10.1371/journal.pone.0089070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stros M, Polanska E, Kucirek M, Pospisilova S. 2015. Histone H1 differentially inhibits DNA bending by reduced and oxidized HMGB1 protein. PLoS One 10:e0138774. doi: 10.1371/journal.pone.0138774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Postnikov YV, Bustin M. 2016. Functional interplay between histone H1 and HMG proteins in chromatin. Biochim Biophys Acta 1859:462–467. doi: 10.1016/j.bbagrm.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cato L, Stott K, Watson M, Thomas JO. 2008. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J Mol Biol 384:1262–1272. doi: 10.1016/j.jmb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 126.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 127.Ray S, Grove A. 2009. The yeast high mobility group protein HMO2, a subunit of the chromatin-remodeling complex INO80, binds DNA ends. Nucleic Acids Res 37:6389–6399. doi: 10.1093/nar/gkp695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lu J, Kobayashi R, Brill SJ. 1996. Characterization of a high mobility group 1/2 homolog in yeast. J Biol Chem 271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- 129.Neuvéglise C, Bon E, Lépingle A, Wincker P, Artiguenave F, Gaillardin C, Casarégola S. 2000. Genomic exploration of the hemiascomycetous yeasts. 9. Saccharomyces kluyveri. FEBS Lett 487:56–60. doi: 10.1016/S0014-5793(00)02280-8. [DOI] [PubMed] [Google Scholar]
- 130.Albert B, Colleran C, Léger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O. 2013. Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res 41:10135–10149. doi: 10.1093/nar/gkt770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kamau E, Bauerle KT, Grove A. 2004. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J Biol Chem 279:55234–55240. doi: 10.1074/jbc.M409459200. [DOI] [PubMed] [Google Scholar]
- 133.Bauerle KT, Kamau E, Grove A. 2006. Interactions between N-and C-terminal domains of the Saccharomyces cerevisiae high-mobility group protein HMO1 are required for DNA bending. Biochemistry 45:3635–3645. doi: 10.1021/bi0522798. [DOI] [PubMed] [Google Scholar]
- 134.Xiao L, Williams AM, Grove A. 2010. The C-terminal domain of yeast high mobility group protein HMO1 mediates lateral protein accretion and in-phase DNA bending. Biochemistry 49:4051–4059. doi: 10.1021/bi1003603. [DOI] [PubMed] [Google Scholar]
- 135.Xiao L, Kamau E, Donze D, Grove A. 2011. Expression of yeast high mobility group protein HMO1 is regulated by TOR signaling. Gene 489:55–62. doi: 10.1016/j.gene.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 136.Dolinski KJ, Heitman J. 1999. Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics 151:935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arevalo-Rodriguez M, Wu X, Hanes SD, Heitman J. 2004. Prolyl isomerases in yeast. Front Biosci 9:2420–2446. doi: 10.2741/1405. [DOI] [PubMed] [Google Scholar]
- 138.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. 2007. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Albert B, Perez-Fernandez J, Léger-Silvestre I, Gadal O. 2012. Regulation of ribosomal RNA production by RNA polymerase I: does elongation come first? Genet Res Int 2012:276948. doi: 10.1155/2012/276948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meraner J, Lechner M, Loidl A, Goralik-Schramel M, Voit R, Grummt I, Loidl P. 2006. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res 34:1798–1806. doi: 10.1093/nar/gkl101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.James A, Wang Y, Raje H, Rosby R, DiMario P. 2014. Nucleolar stress with and without p53. Nucleus 5:402–426. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsai RY, Pederson T. 2014. Connecting the nucleolus to the cell cycle and human disease. FASEB J 28:3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- 144.Huang Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res 30:1465–1482. doi: 10.1093/nar/30.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wittner M, Hamperl S, Stöckl U, Seufert W, Tschochner H, Milkereit P, Griesenbeck J. 2011. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell 145:543–554. doi: 10.1016/j.cell.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 146.Ryu H-Y, Ahn SH. 2014. Yeast histone H3 lysine 4 demethylase Jhd2 regulates mitotic ribosomal DNA condensation. BMC Biol 12:75. doi: 10.1186/s12915-014-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Srivastava R, Srivastava R, Ahn SH. 2016. The epigenetic pathways to ribosomal DNA silencing. Microbiol Mol Biol Rev 80:545–563. doi: 10.1128/MMBR.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. 2008. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev 22:1190–1204. doi: 10.1101/gad.466908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hall DB, Wade JT, Struhl K. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol 26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Berger AB, Decourty L, Badis G, Nehrbass U, Jacquier A, Gadal O. 2007. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol Cell Biol 27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Murugesapillai D, McCauley MJ, Huo R, Nelson Holte MH, Stepanyants A, Maher LJ III, Israeloff NE, Williams MC. 2014. DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Res 42:8996–9004. doi: 10.1093/nar/gku635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tremblay M, Charton R, Wittner M, Levasseur G, Griesenbeck J, Conconi A. 2014. UV light-induced DNA lesions cause dissociation of yeast RNA polymerases-I and establishment of a specialized chromatin structure at rRNA genes. Nucleic Acids Res 42:380–395. doi: 10.1093/nar/gkt871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsang CK, Li H, Zheng XS. 2007. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J 26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang D, Mansisidor A, Prabhakar G, Hochwagen A. 2016. Condensin and Hmo1 mediate a starvation-induced transcriptional position effect within the ribosomal DNA array. Cell Rep 14:1010–1017. doi: 10.1016/j.celrep.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gadal O, Labarre S, Boschiero C, Thuriaux P. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J 21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. 2008. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol 28:1596–1605. doi: 10.1128/MCB.01464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Albert B, Leger-Silvestre I, Normand C, Ostermaier MK, Perez-Fernandez J, Panov KI, Zomerdijk JC, Schultz P, Gadal O. 2011. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J Cell Biol 192:277–293. doi: 10.1083/jcb.201006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kuhn A, Grummt I. 1992. Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc Natl Acad Sci U S A 89:7340–7344. doi: 10.1073/pnas.89.16.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tuan JC, Zhai W, Comai L. 1999. Recruitment of TATA-binding protein–TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol Cell Biol 19:2872–2879. doi: 10.1128/MCB.19.4.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Voit R, Grummt I. 2001. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci U S A 98:13631–13636. doi: 10.1073/pnas.231071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O'Sullivan AC, Sullivan GJ, McStay B. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22:657–658. doi: 10.1128/MCB.22.2.657-668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Copenhaver GP, Putnam CD, Denton ML, Pikaard CS. 1994. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res 22:2651–2657. doi: 10.1093/nar/22.13.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wilson DN, Cate JHD. 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 4:a011536. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li B, Nierras CR, Warner JR. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol 19:5393–5404. doi: 10.1128/MCB.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu L, Morse RH. 1999. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol 19:5279–5288. doi: 10.1128/MCB.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kasahara K, Ohyama Y, Kokubo T. 2011. Hmo1 directs pre-initiation complex assembly to an appropriate site on its target gene promoters by masking a nucleosome-free region. Nucleic Acids Res 39:4136–4150. doi: 10.1093/nar/gkq1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kasahara K, Ki S, Aoyama K, Takahashi H, Kokubo T. 2008. Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II. Nucleic Acids Res 36:1343–1357. doi: 10.1093/nar/gkm1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kotani T, Miyake T, Tsukihashi Y, Hinnebusch AG, Nakatani Y, Kawaichi M, Kokubo T. 1998. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J Biol Chem 273:32254–32264. doi: 10.1074/jbc.273.48.32254. [DOI] [PubMed] [Google Scholar]
- 170.Knight B, Kubik S, Ghosh B, Bruzzone MJ, Geertz M, Martin V, Dénervaud N, Jacquet P, Ozkan B, Rougemont J. 2014. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev 28:1695–1709. doi: 10.1101/gad.244434.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 172.Rudra D, Zhao Y, Warner JR. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 174.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A 101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Reja R, Vinayachandran V, Ghosh S, Pugh BF. 2015. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev 29:1942–1954. doi: 10.1101/gad.268896.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Panday A, Xiao L, Grove A. 2015. Yeast high mobility group protein HMO1 stabilizes chromatin and is evicted during repair of DNA double strand breaks. Nucleic Acids Res 43:5759–5770. doi: 10.1093/nar/gkv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 179.Hepp MI, Alarcon V, Dutta A, Workman JL, Gutierrez JL. 2014. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim Biophys Acta 1839:764–772. doi: 10.1016/j.bbagrm.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shimobayashi M, Hall MN. 2016. Multiple amino acid sensing inputs to mTORC1. Cell Res 26:7–20. doi: 10.1038/cr.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Workman JJ, Chen H, Laribee RN. 2014. Environmental signaling through the mechanistic target of rapamycin complex 1: mTORC1 goes nuclear. Cell Cycle 13:714–725. doi: 10.4161/cc.28112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Betz C, Hall MN. 2013. Where is mTOR and what is it doing there? J Cell Biol 203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wullschleger S, Loewith R, Oppliger W, Hall MN. 2005. Molecular organization of target of rapamycin complex 2. J Biol Chem 280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 184.Tsang CK, Bertram PG, Ai W, Drenan R, Zheng XS. 2003. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J 22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 186.Chen H, Workman JJ, Tenga A, Laribee RN. 2013. Target of rapamycin signaling regulates high mobility group protein association to chromatin, which functions to suppress necrotic cell death. Epigenetics Chromatin 6:29. doi: 10.1186/1756-8935-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen H, Fan M, Pfeffer LM, Laribee RN. 2012. The histone H3 lysine 56 acetylation pathway is regulated by target of rapamycin (TOR) signaling and functions directly in ribosomal RNA biogenesis. Nucleic Acids Res 40:6534–6546. doi: 10.1093/nar/gks345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Williams SK, Truong D, Tyler JK. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci U S A 105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wade JT, Hall DB, Struhl K. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- 190.Rudra D, Mallick J, Zhao Y, Warner JR. 2007. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol 27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xiao L, Grove A. 2009. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr Genomics 10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Usdin K, House NC, Freudenreich CH. 2015. Repeat instability during DNA repair: insights from model systems. Crit Rev Biochem Mol Biol 50:142–167. doi: 10.3109/10409238.2014.999192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim H, Livingston DM. 2006. A high mobility group protein binds to long CAG repeat tracts and establishes their chromatin organization in Saccharomyces cerevisiae. J Biol Chem 281:15735–15740. doi: 10.1074/jbc.M512816200. [DOI] [PubMed] [Google Scholar]
- 194.Aguilera A, Gomez-Gonzalez B. 2008. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 195.Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K. 2009. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 196.Ghosal G, Chen J. 2013. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res 2:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gonzalez-Huici V, Szakal B, Urulangodi M, Psakhye I, Castellucci F, Menolfi D, Rajakumara E, Fumasoni M, Bermejo R, Jentsch S, Branzei D. 2014. DNA bending facilitates the error-free DNA damage tolerance pathway and upholds genome integrity. EMBO J 33:327–340. doi: 10.1002/embj.201387425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Panday A, Grove A. 2016. The high mobility group protein HMO1 functions as a linker histone in yeast. Epigenetics Chromatin 9:1. doi: 10.1186/s13072-015-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gursoy-Yuzugullu O, House N, Price BD. 2016. Patching broken DNA: nucleosome dynamics and the repair of DNA breaks. J Mol Biol 428:1846–1860. doi: 10.1016/j.jmb.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pongpanich M, Patchsung M, Thongsroy J, Mutirangura A. 2014. Characteristics of replication-independent endogenous double-strand breaks in Saccharomyces cerevisiae. BMC Genomics 15:750. doi: 10.1186/1471-2164-15-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Thongsroy J, Matangkasombut O, Thongnak A, Rattanatanyong P, Jirawatnotai S, Mutirangura A. 2013. Replication-independent endogenous DNA double-strand breaks in Saccharomyces cerevisiae model. PLoS One 8:e72706. doi: 10.1371/journal.pone.0072706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Grove A. 2011. Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol 13:1–12. doi: 10.21775/cimb.013.001. [DOI] [PubMed] [Google Scholar]