FIG 3.
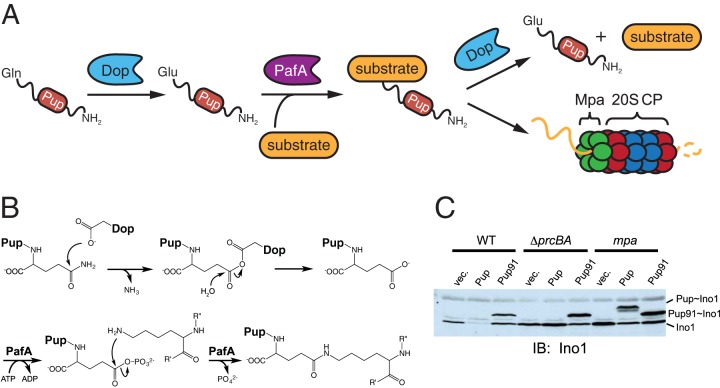
ATP-dependent degradation requires pupylation. (A) From left to right, Pup is deamidated at its C terminus by Dop, producing PupGlu; PafA ligates PupGlu to a substrate; and a pupylated substrate can either be depupylated by Dop or be degraded by the proteasome. (B) Proposed mechanisms of deamidation and pupylation. Starting from top left, the Pup C-terminal Gln is attacked by a nucleophilic Asp in Dop, forming a Pup-Dop intermediate; Pup undergoes a nucleophilic attack by water, activated by Dop, to yield PupGlu; PafA uses ATP to phosphorylate the γ-carboxylate of PupGlu; and nucleophilic attack by a substrate Lys residue results in the covalent linkage of Pup to a substrate. Another mechanism of Pup deamidation has been proposed, in which the Dop Asp residue coordinates a water molecule and nucleophilic attack by this activated water results in deamidation (66). (C) Depupylation of a pupylated substrate in M. smegmatis requires an intact Pup N terminus as well as Mpa. The top labels indicate the M. smegmatis strain background, as follows: WT, wild-type strain; ΔprcBA, deletion mutant lacking the prcBA genes; and mpa, a strain in which the gene has been disrupted with an integrated plasmid. Lanes: vec., empty vector; Pup, overproduced full-length Pup; Pup91, overproduced truncated Pup missing 30 N-terminal amino acids. Immunoblotting (IB) was performed on cell lysates from the indicated strains by using an antibody to M. tuberculosis Ino1 (inositol-1-phosphate synthetase), a known pupylated substrate. Bands corresponding to Ino1 and its Pup or Pup91 conjugate are indicated at the right. (Reprinted from reference 50 with permission from Elsevier.)