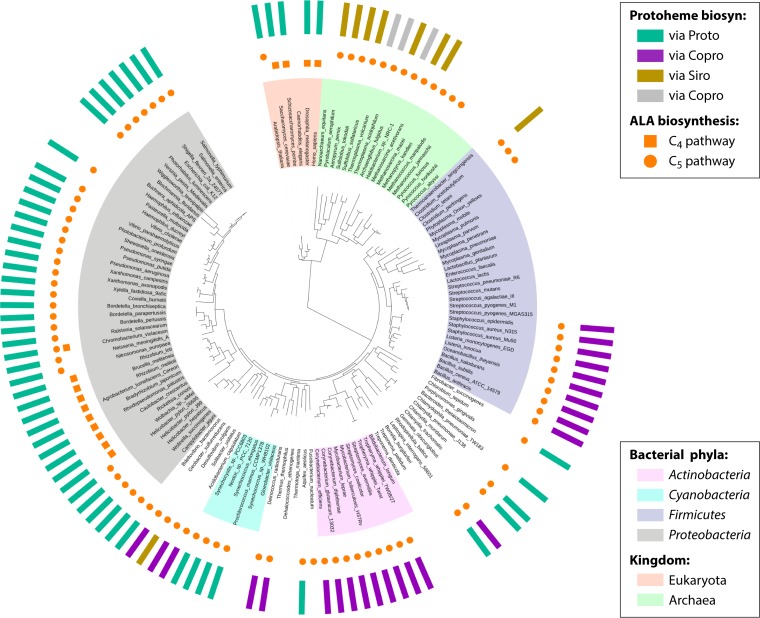
FIG 21.
Phylogenic distribution of the three currently characterized pathways for protoheme synthesis mapped onto the Tree of Life. The outside ring shows (as vertical rectangles) the presence or absence of the siroheme-dependent branch (via Siro), the coproporphyrin-dependent branch (via Copro), or the classic protoporphyrin-dependent branch (via Proto). Gray rectangles mark the organisms that contain unusual combinations of genes normally involved in different pathways for protoheme synthesis (hybrid paths). The distribution of the two routes used to synthesize 5-aminolevulinic acid (ALA) are also presented: the Shemin or C4 pathway and the C5 pathway. The absence of a circle or square shows the inability of an organism to produce tetrapyrroles of any kind. Likewise, the absence of a rectangle in the outside ring indicates the absence of any known route for protoheme synthesis in an organism. This illustration covers only 133 representative organisms (17 archaea, 110 eubacteria, and 6 eukaryotes) included in the Tree of Life (391, 392). A full analysis of the 978 representative microorganisms performed in this work is available in the SEED subsystem “Heme Biosynthesis: Protoporphyrin-, Coproporphyrin-, and Siroheme-Dependent Pathways” (see http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowFunctionalRoles&subsystem=Heme_Biosynthesis%3A_protoporphyrin-%2C_coproporphyrin-_and_siroheme-dependent_pathways). Mapped onto this tree, the siroheme-dependent pathway occurs largely in the Archaea, in the Thermodesulfobacteria, and, rarely, in several other taxa (see Table S3 in the supplemental material). The CPD route (teal) is found primarily in slow-evolving monoderm Firmicutes and Actinobacteria and in evolutionarily early-branching (Acidobacteria, Planctomyces, and Aquificae) and transitional (Deinococcus-Thermus group) diderm phyla, while the PPD pathway has a wider distribution, occurring in Proteobacteria, cyanobacteria, the Bacteroidetes-Chlorobi and Chlamydiae-Verrucomicrobia groups, Aquificae, Gemmatimonadetes, Caldithrix, and several other taxa. This is the main route for protoheme production in the evolutionarily younger Proteobacteria, with an illuminating exception of the Deltaproteobacteria. Deltaproteobacteria and epsilonproteobacteria are believed to be the oldest phyla within the Proteobacteria, with the other main proteobacterial groups being derived from them linearly in a directional rather than in a tree-like manner in the order delta/epsilonproteobacteria → alphaproteobacteria → betaproteobacteria → gammaproteobacteria (393). Notably, the phylum Deltaproteobacteria is the only proteobacterial phylum in which all 3 routes leading to protoheme are represented in different species (Table S3). A complete reconstruction of the heme biosynthetic pathways in the 38 representative deltaproteobacterial genomes is available online in the SEED subsystem (limit view to “deltaproteobacteria”). As a few examples, genomes of Stigmatella aurantiaca and Myxococcus xanthus encode the PPD pathway, and genomes of Geobacter metallireducens and Desulfuromonas acetoxidans harbor the CPD pathway, while genomes of Desulfovibrio vulgaris, Desulfatibacillum alkenivorans, and Desulfobacula toluolica harbor the siroheme-dependent route.