Figure 3.
Lineage Analysis of HuCNS-SC CCL and RCL in the 60 DPI Paradigm
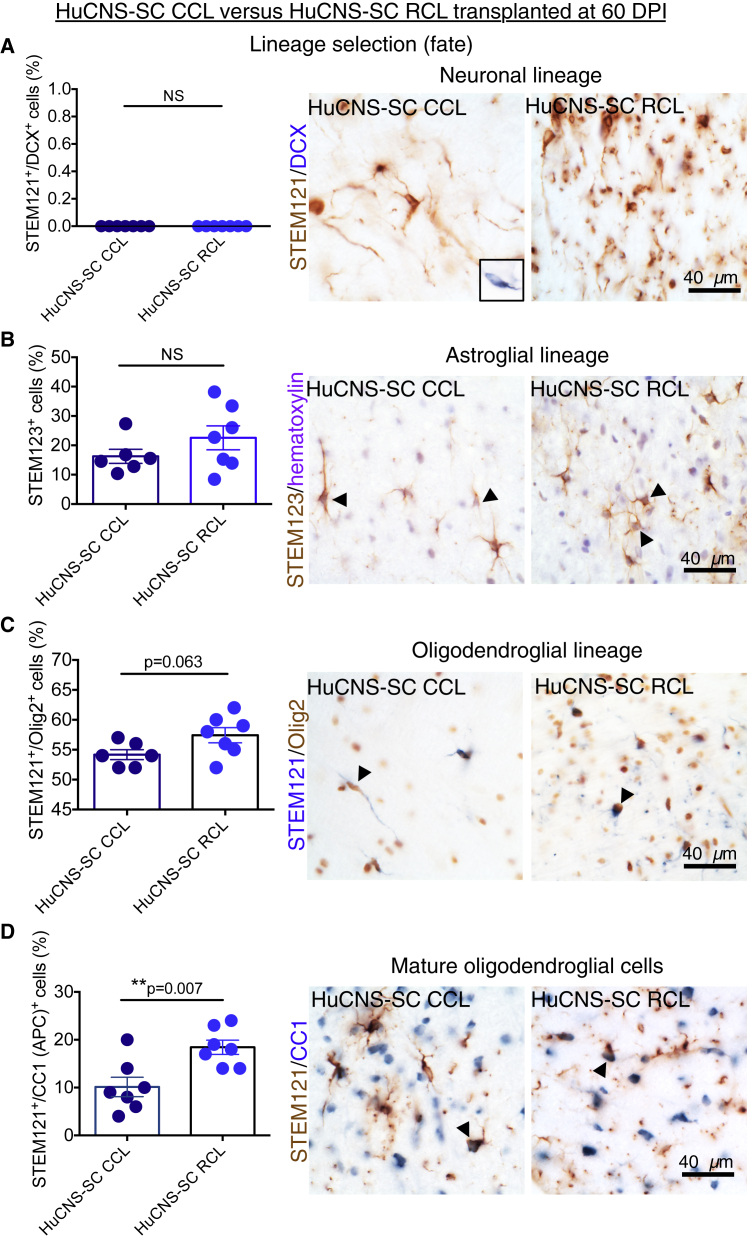
Data for proportional human cell fate were collected by blinded, unbiased stereology.
(A) Co-immunostaining for STEM121 (brown) and the early neuronal marker DCX (blue) revealed no STEM121+/DCX+ cells in either the CCL (n = 7) or RCL (n = 7) groups (Student's two-tailed t test, p > 0.05, n.s.). Inset shows positive control for DCX in the hippocampus.
(B) Immunostaining for human-specific GFAP (STEM123, brown) with hematoxylin counterstaining (purple) revealed no significant difference in STEM123 proportion between the CCL (n = 6) and RCL (n = 7) groups (Student's two-tailed t test, p > 0.2).
(C) Immunostaining for STEM121 (blue) and the oligodendroglial nuclear marker Olig2 (brown) revealed that the largest proportion of STEM121+ cells were also nuclear Olig2+ and there was a trend for a decrease in Olgi2+ cells in CCL (n = 5) versus RCL (n = 7) transplants (Student's two-tailed t test, p = 0.06).
(D) Immunostaining for STEM121 (brown) and the mature oligodendroglial marker CC1 (blue) revealed a significant decrease in the proportion of STEM121+/CC1+ cells in CCL (n = 7) versus RCL (n = 7) transplants (Student's two-tailed t test, p < 0.007).
Arrowheads indicate double-positive cells. Data shown as means ± SEM.