Summary
Metabolism of oxidative stress is necessary for cellular survival. We have previously utilized the zebrafish as a model of the oxidative stress response. In this study, we found that gata1-expressing erythroid cells contributed to a significant proportion of total-body oxidative stress when animals were exposed to a strong pro-oxidant. RNA-seq of zebrafish under oxidative stress revealed the induction of tp53. Zebrafish carrying tp53 with a mutation in its DNA-binding domain were acutely sensitive to pro-oxidant exposure and displayed significant reactive oxygen species (ROS) and tp53-independent erythroid cell death resulting in an edematous phenotype. We found that a major contributing factor to ROS was increased basal mitochondrial respiratory rate without reserve. These data add to the concept that tp53, while classically a tumor suppressor and cell-cycle regulator, has additional roles in controlling cellular oxidative stress.
Keywords: erythroid precursors, oxidative stress, tp53, zebrafish, reactive oxygen species, mitochondria
Highlights
-
•
Erythroid precursors contribute significantly to total ROS after oxidative challenge
-
•
Tp53 is induced after pro-oxidant challenge
-
•
Mutated tp53 is associated with an increased mitochondrial oxygen consumption rate
-
•
Decreased mitochondrial reserve leads to overwhelming ROS and erythroid cell death
In this article, Lund and colleagues show that Gata1+ erythroid precursors contribute to total reactive oxygen species (ROS) in the zebrafish. Disruption of Tp53 increases ROS and overall sensitivity to pro-oxidant challenge, involving dysregulation of mitochondrial respiration.
Introduction
Cells undergoing oxidative respiration rid the cell of undesirable metabolites, such as reactive oxygen species (ROS), to avoid oxidative stress and cellular damage. Most cells have a specific cellular response when exposed to compounds (toxins, medicines, foods) that increase free radical production beyond what can be detoxified. Recent data suggest that the oxidative stress response can involve the TP53 pathway where TP53 can function both as a sensor of ROS and ROS-mediated DNA damage but may also be involved in the regulation of ROS levels (reviewed by Sharpless and DePinho, 2002). In some situations, TP53 can function as an antioxidant, given that when TP53 levels were reduced via small interfering RNA (siRNA) in a variety of cell lines, oxidative stress and ROS increased 2-fold (Sablina et al., 2005). Specific target genes implicated as part of the antioxidant pathway mediated by TP53 included SESN2, SESN1, GPX1, and CDKN1A (Budanov and Karin, 2008, Sablina et al., 2005). Alternatively, TP53 can stabilize the anti-apoptotic protein, BCLXL, at the mitochondrial outer membrane. In the absence of TP53, the mitochondrial outer membrane is destabilized and cytochrome c is released (Mihara et al., 2003). Cell death then occurs through the “mitochondrial pathway,” now recently termed necroptosis (Jouan-Lanhouet et al., 2014).
Mature red cells are exposed to both extrinsic and intrinsic ROS, which can lead to impairment of membrane deformability, reduced red cell lifespan, and reduced oxygen delivery (Mohanty et al., 2014, Perrone et al., 2012). Very little is known about how erythroid progenitors process oxidative stress, although mechanisms are likely different, given that they retain both nuclei and mitochondria, which mediate metabolism distinct from their progeny. We have previously used the zebrafish to model oxidative stress in Gata1+ erythroid cells caused by glucose-6-phosphate dehydrogenase (G6PD) deficiency (Patrinostro et al., 2013). Gata1+ erythroid cells with reduced G6PD activity developed elevated levels of ROS and were very sensitive to cell lysis with pro-oxidant exposure. Specific mechanisms of how Gata1+ erythroid cells respond to oxidative stress remain unknown. The zebrafish is an excellent model of hemato- and erythropoiesis and has many of the conserved genetic regulators of hematopoiesis, including gata1, lck, and c-myb (Bahary and Zon, 1998, Davidson and Zon, 2004). There have been several models of human erythroid disorders created in the zebrafish, including Diamond-Blackfan anemia, porphyria, and hereditary spherocytosis (Dooley et al., 2008, Taylor et al., 2012).
In this article, we describe the effects of pro-oxidant exposure on Gata1+ erythroid cells. Specifically, we found that Gata1+ erythroid cells are a significant source of total-body ROS after pro-oxidant exposure in zebrafish early in development. Furthermore, we determined that a specific program associated with tp53 activation drives the response to pro-oxidant exposure, and mutation in tp53 was associated with increased basal mitochondrial respiration to maximal levels. This created a situation of decreased mitochondrial respiratory capacity when encountering a pro-oxidant challenge and elevated ROS, resulting in increased cell death.
Results
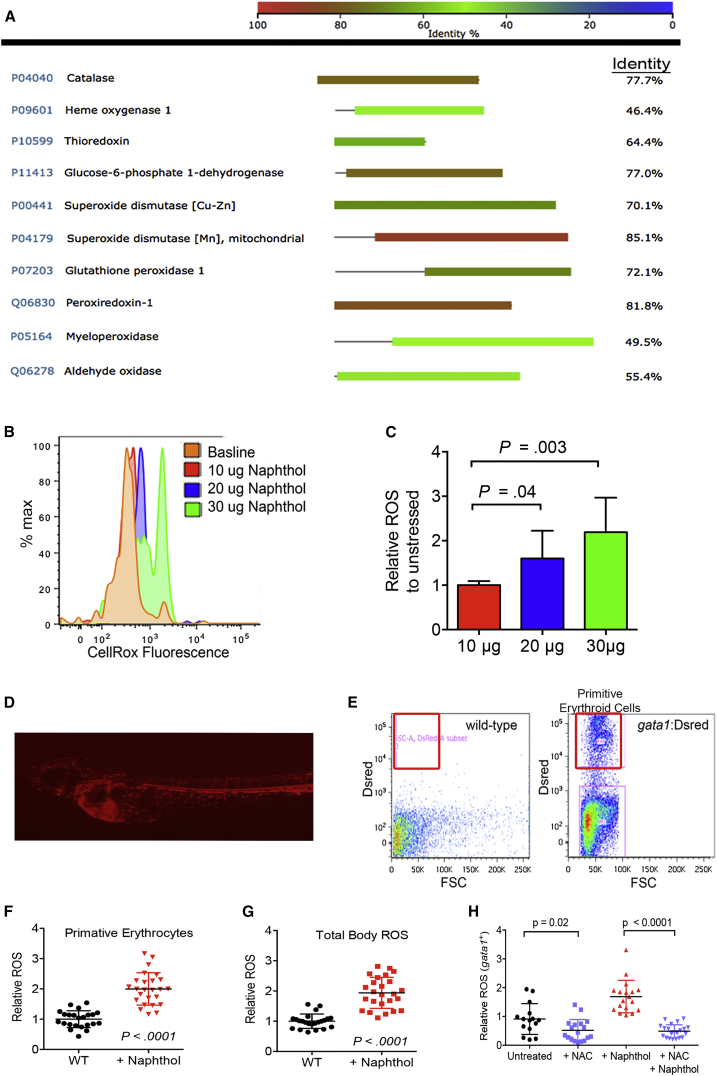
We previously published a zebrafish model of g6pd deficiency that showed acute sensitivity to pro-oxidant challenge with robust ROS generation and significant erythroid cell death when tested in developing zebrafish (Patrinostro et al., 2013). The zebrafish is an advantageous model to study oxidative stress, as many key proteins involved in reduction-oxidation (redox) and the response to oxidative stress are conserved in sequence and function. Figure 1A shows amino acid identity between human and zebrafish oxidative stress response proteins ranging from 46.4% in heme oxygenase-1 to 85.5% in mitochondrial superoxide dismutase. Considering conserved amino acid substitutions, the homologies between these human and zebrafish proteins are close to 90% (not shown), allowing the zebrafish to serve as a robust model of oxidative stress.
Figure 1.
Oxidative Stress in gata1+ Erythroid Cells
(A) BLASTP identity analysis of key proteins involved in the oxidative stress response between Danio rerio and Homo sapiens.
(B) Representative flow cytometry histograms showing CellRox emission in 72 hpf embryos after 48 hr of 1-naphthol exposure (concentrations shown as μg per 5 mL).
(C) Quantification of CellRox probe signal MFI relative to unstressed embryos. n = 20 individual animals per condition showing one of five independent experiments.
(D) Live imaging of a gata1:DsRed transgenic zebrafish at 72 hpf indicating DsRed-positive primitive erythroid cells.
(E) Representative flow cytometry of single-cell suspensions prepared from wild-type and gata1:DsRed zebrafish at 72 hpf.
(F and G) Pro-oxidant exposure induces ROS in Gata1+ erythroid precursors. Animals were exposed to 20 μg/5 mL 1-naphthol followed by flow cytometry of Gata1+ erythroid cells by gating on DsRed-positive cells (or all cells for total-body ROS) and measuring CellRox probe MFI to determine ROS. n = 20–25 individual animals per condition showing one of four independent experiments.
(H) NAC combined with 1-naphthol reduces ROS in Gata1+ erythroid cells. All pro-oxidant exposure times were from 24 to 72 hpf.
All data are shown as the mean ± SD, with the p value from a Student t test. See also Figure S1.
To quantify the amount of total ROS generated with pro-oxidant exposure, we exposed wild-type animals at 24 hr postfertilization (hpf) to increasing doses of the prototypical pro-oxidant, 1-naphthol, for 48 hr, as we previously described (Patrinostro et al., 2013). Using the fluorescent indicator probe, CellRox, to indicate total-body ROS generation (Patrinostro et al., 2013), we found a measurable dose-response increase in ROS levels in response to pro-oxidant challenge (Figures 1B and 1C). A specific benefit to using zebrafish as a model is the ability to efficiently create animals with fluorescently marked cell types. We utilized the gata1:DsRed zebrafish, which has DsRed-expressing Gata-1+ erythroid precursors that can be visualized microscopically as well as localized by flow cytometry (Figures 1D and 1E) (Traver et al., 2003). Pro-oxidant exposure induced ROS in Gata1+ erythroid cells to a similar magnitude as that of the total-body ROS (p < 0.0001, Figures 1F and 1G). The addition of the potent antioxidant, n-acetylcysteine (NAC), to the water was able to efficiently rescue animals from significant erythroid ROS production (Figure 1H).
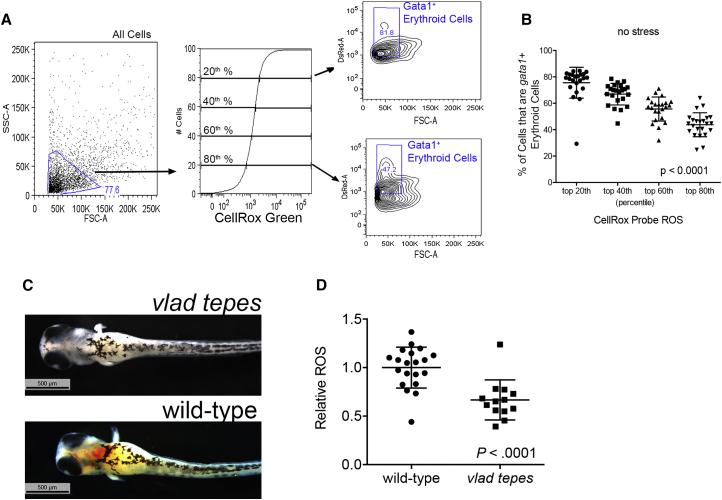
To validate the contribution of Gata1+ erythroid cells to total-body ROS, we evaluated the total CellRox probe fluorescence intensity and divided the intensity into quintiles. By determining the proportion of Gata1+ cells in each quintile, we determined that a large fraction of the highest ROS was produced by erythroid cells (Figures 2A and 2B). To confirm that erythroid cells produced elevated levels of ROS, we utilized the Vlad tepes zebrafish, which has a mutation in the gata1 gene and does not develop Gata1+ erythroid cells (primitive or definitive), although is still able to survive for the first 10 days of embryonic life (Figure 2C) (Lyons et al., 2002). Total-body pro-oxidant-induced ROS generation in Vlad tepes was 50% reduced when compared with wild-type animals (p < 0.0001, Figure 2D). Collectively, these data show that Gata1+ erythroid cells have measurable and robust ROS after pro-oxidant challenge and are a significant source of total-body ROS.
Figure 2.
Gata1+ Erythroid Cells Contribute a Significant Proportion of ROS to Total-Body ROS
(A) Gata1+ primitive erythroid cells contribute to total ROS. Cells from stressed gata1:DsRed zebrafish were gated initially for the total cell population, then evaluated by cumulative distribution function of CellROX green ROS probe signal. The 20th to 80th percentiles of ROS signal were gated, and the makeup of each percentile in terms of the gata1:DsRed fraction of all cells was determined. n = 20–25 individual animals per condition showing one of three independent experiments.
(B) Makeup of each percentile of ROS probe signal in terms of % gata1:DsRed contribution showing that the highest ROS signal originates mostly from Gata1+ erythroid cells. One-way ANOVA indicates p value < 0.0001 for overall effect and p < 0.001 in two-way comparisons between all groups.
(C) o-Dianisidine-stained vlad tepes zebrafish at 72 hpf showing a lack of erythrocytes. Scale bars represent 500 μm.
(D) Vlad tepes zebrafish have reduced total-body ROS. Vlad tepes and phenotypically normal clutch mates were treated with 20 μg/5 mL 1-naphthol followed by flow cytometry of total-body ROS indicated by CellRox ROS probe (n = 17–22 individual animals per condition in one of two experiments). All pro-oxidant exposure times were from 24 to 72 hpf.
All data are shown as the mean ± SD, with the p value from a Student t test, unless otherwise noted.
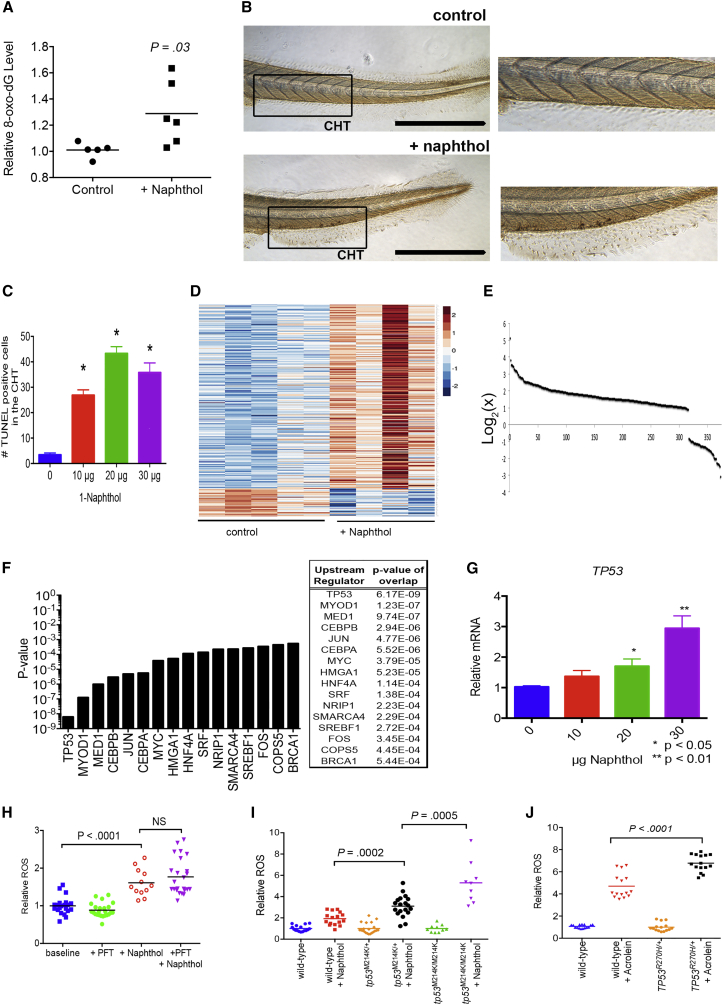
Exposure to pro-oxidant challenge produces several molecular modifications, including protein carbonylation and lipid peroxidation, and induces DNA toxicity through deleterious modification (Valko et al., 2007). To explore potential DNA-toxic effects of an external pro-oxidant challenge, we measured the concentration of 8-oxo-2ʹ-deoxyguanosine (8-oxo-dG) in zebrafish after pro-oxidant exposure; 8-oxo-dG is an oxidized derivative of deoxyguanosine and a marker of DNA damage from oxidative stress (Delaney et al., 2012). We found that levels of 8-oxo-dG increased by 30% (p = 0.03) in zebrafish embryos after 48 hr of pro-oxidant exposure (Figure 3A). As an additional measure of DNA damage, we performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays. TUNEL assay indicated that DNA damage was occurring both in circulating cells (which are mostly primitive Gata1+ erythroid cells at 72 hpf development) (Davidson and Zon, 2004), as well as in the caudal hematopoietic tissue (CHT), the anatomic area where definitive hematopoiesis takes place, shown in Figures 3B and 3C (Bertrand et al., 2007). Acridine orange staining of stressed embryos also indicated that cell death was occurring in many Gata1+ cells of the CHT (Figure S2).
Figure 3.
Oxidative Stress Induces tp53, which Can Modulate ROS Levels
(A) Zebrafish show increased 8-oxo-dG after pro-oxidant exposure (1-naphthol at 20 μg/5 mL). n = 5–6 per condition, one of two independent experiments.
(B) Representative image of a TUNEL assay from zebrafish after pro-oxidant exposure (20 μg/5 mL). Boxed region indicates the CHT. Scale bar represents 500 μm.
(C) Number of TUNEL-positive cells present in the CHT. n = 14–16 individual animals per condition in one of two independent experiments. ∗p < 0.0001.
(D) Heatmap of RNA-seq data from untreated and 1-naphthol-treated zebrafish. Unsupervised clustering was performed on genes with log2 expression >2-fold difference.
(E) Distribution of genes with log2 expression >2-fold difference between control and 1-naphthol-treated zebrafish. n = 5 control animals and n = 6 naphthol-treated animals.
(F) IPA results of the significant Upstream Regulators having greater expression in 1-naphthol-treated zebrafish.
(G) RT-PCR of tp53 in zebrafish after pro-oxidant exposure (n = 20 individual animals per group, technical duplicates, pooled from four independent experiments). ∗p < 0.05, ∗∗p < 0.01.
(H) Tp53 inactivation using pifithrin (PFT) during pro-oxidant exposure (1-naphthol at 30 μg/5 mL) at 1 μM final concentration increases ROS in Gata1+-expressing erythrocytes. ROS measured using the CellRox ROS probe relative to untreated group. n = 20–25 individual animals per condition showing one of three independent experiments. See also Figure S2G.
(I) Elevated ROS in tp53M214K zebrafish Gata1+-expressing erythroid cells after pro-oxidant challenge. n = 20–25 individual animals per condition showing one of three independent experiments.
(J) Erythroid progenitors in TP53R270H/+murine whole bone marrow have increased ROS generation after short-term oxidative challenge. After exposure to 1 mM acrolein, nucleated CD71+ cells were gated by flow cytometry (see Figure S2H for gating) and ROS measured using CellROX green probe. ROS is given relative to the untreated group. Shown are data pooled from two independent experiments. All zebrafish pro-oxidant exposure times were from 24 to 72 hpf. n = 10–20 individual animals per condition pooled from two independent experiments.
All data are shown as the mean ± SD, with the p value from a Student t test.
To learn what specific genes and biological pathways are involved in the response to pro-oxidant exposure, we performed RNA-seq analysis on 72-hpf embryos after 48 hr of pro-oxidant exposure; the heatmap analysis of unsupervised clustering is shown in Figure 3D. We found 316 genes upregulated (increased >2-fold, Table S1) in response to pro-oxidant exposure and 56 genes downregulated (decreased >2-fold), as shown in Figure 3E. To determine if there was a common regulatory pathway governing the pro-oxidant-induced genes, we performed Ingenuity Pathway Analysis (IPA; Ingenuity Systems) to identify networks of specific upstream transcription factor regulators and found that the tp53 pathway was the most significant regulator involved in the oxidative stress response (p = 6.17 × 10−9, Figure 3F). This was not surprising given our prior findings of DNA modification and DNA damage that occurred after pro-oxidant exposure. We performed qRT-PCR to determine if tp53 expression was changed after 1-naphthol exposure and found a 2- to 3-fold induction at the highest amounts of pro-oxidant (Figure 3G). In addition, qRT-PCR of fluorescence-activated cell sorted stressed Gata1+ erythroid cells showed induction of tp53 as well as known downstream tp53 targets, including sestrin 2 (sens2), nuclear Factor Erythroid 2-Related Factor 2 (nrf2), heme oxygenase 1 (hmox), and thioredoxin (txn) that are upregulated by ROS (Figures S2C–S2F).
To study the effect of tp53 inhibition and oxidative stress, we utilized pifithrin-alpha, a small molecule inhibitor of TP53 activity (Duffy and Wickstrom, 2007, Rocha et al., 2003). Pifithrin is thought to block the transcription factor activity of TP53, as well as protein-protein interactions, and is able to protect cells from TP53-dependent apoptosis (Culmsee et al., 2001). We found that exposure to pifithrin from 24 to 72 hpf led to a slight (but not significant) increase in ROS generated by Gata1+ erythroid cells during simultaneous pro-oxidant exposure (p = 0.50, Figure 3H). Given the relatively short half-life of pifithrin (approximately 4 hr), it is possible that Tp53 was not sufficiently inhibited (Gary and Jensen, 2005), although exposure of stressed embryos to pulses of pifithrin every 4 hr also did not significantly alter ROS (Figure S2G). Therefore, we decided to employ an alternative approach using a well-characterized zebrafish with a tp53 mutation that disables Tp53 DNA-binding activity. The tp53M214K zebrafish has a tp53 mutation (methionine-214 to lysine, M214K) known to be analogous to the human TP53 M246K mutation commonly found in human cancers (Berghmans et al., 2005). The M214K mutation disrupts the DNA-binding domain of Tp53, rendering it unable to upregulate p21 after UV irradiation and animals also show cellular resistance to apoptosis. Furthermore, zebrafish harboring this mutation develop malignancies early in life (starting at 8 months of age) (Berghmans et al., 2005). We crossed tp53M214K animals to the gata1:DsRed transgenic line, followed by in-crossing to generate offspring in which tp53 was mutated in DsRed-positive Gata1+ erythroid cells. Pro-oxidant exposure of tp53M214K heterozygous zebrafish generated 3-fold higher levels of ROS in Gata1+ erythroid cells, in comparison with wild-type counterparts (p = 0.0002, Figure 3I). At the same time, total-body ROS level was also significantly increased (data not shown). Similarly, using tp53M214K homozygous animals, the ROS levels increased 5-fold above that of wild-type (p < 0.0001).
To understand if TP53 played a similar role in mammalian erythroid cells, we evaluated ROS production in erythroid precursors from wild-type and Tp53R270H/+ mice. Tp53R270H/+ mice harbor the Tp53 mutation arginine-270 to histidine (R270H), which abolishes TP53 DNA-binding activity (Olive et al., 2004). Like the tp53M214K zebrafish, Tp53R270H/+ mice develop tumors early in life, and their thymocytes show partial resistance to gamma-induced apoptosis (Olive et al., 2004). While zebrafish and other teleosts are uniquely sensitive to 1-naphthol, we used acrolein (a metabolite of cyclophosphamide) as a strong pro-oxidant to induce ROS in mammalian cells. We identified erythroid precursors using antibodies to the transferrin receptor (CD71) and gating on marrow cells with the highest CD71 expression; we ensured that cells would be at an immature stage (erythroblasts), as opposed to mature erythrocytes that have dim to zero CD71 expression (Chen et al., 2009, Marsee et al., 2010, Peslak et al., 2012). Analysis of nucleated CD71+ erythroid precursors after short-term (5 hr) acrolein exposure indicated significantly higher levels were generated in erythroid precursors from TP53R270H/+mice (Figures 3J and S2H, p < 0.0001). These data indicate that TP53 plays a role in the metabolism of oxidative stress and that haploinsufficiency of TP53 allows for increased ROS generation in mammalian and zebrafish erythroid precursors. In zebrafish, homozygous tp53 disruption increased ROS to an even greater extent.
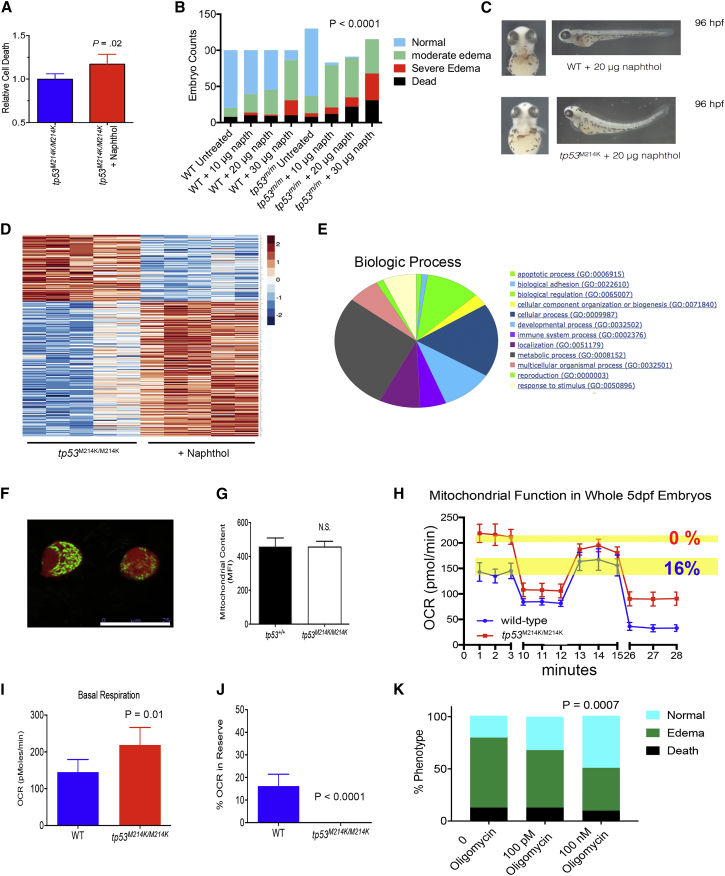
We assessed levels of cell death using annexin V and propidium iodide staining. The tp53M214K/M214K animals showed a 20% increase in erythroid cell death when assessed by propidium iodide staining (p = 0.02, Figures 4A and S3A), but no increase in annexin V staining (Figures S3A and S3B), similar to what was observed with UV irradiation exposure in tp53M214K/M214K animals (Berghmans et al., 2005). These observations suggest that perhaps non-apoptotic pathways were at work, such as necrosis/necroptosis, as previously described (Galluzzi and Kroemer, 2008). When we assessed the phenotype of tp53M214K homozygotes after pro-oxidant exposure, we found a severe reduction in hemoglobin-producing cells shown by o-dianisidine staining. This resulted in a significant amount of edema and increased mortality, especially at higher concentrations of pro-oxidant exposure (Figures 4B and 4C, p < 0.0001). This phenotype was similar to observations we have seen in G6PD-deficient zebrafish under elevated oxidative stress (Patrinostro et al., 2013). Incidentally, pro-oxidant-stressed tp53 mutant zebrafish showed a partial rescue in phenotype when co-exposed with necrostatin, a potent inhibitor of receptor-interacting protein kinase-1 (RIPK1), a central mediator of necroptosis (Degterev et al., 2008), although a more thorough interrogation of necroptotic pathways, such RIP3, would be definitive (Figure S3C).
Figure 4.
Genetic Inactivation of tp53 Leads to Elevated ROS, Cell Death, and Increased Mitochondrial Basal OCR
(A) Relative erythroid cell death indicated by propidium iodide staining cells after 1-naphthol exposure (20 μg/5 mL) and quantified by flow cytometry. n = 20–21 individual animals per group pooled from two independent experiments.
(B) Pro-oxidant exposure (0–30 μg/5 mL) leads to increased death and edema in tp53M214K/M214K zebrafish (mutants denoted as tp53m/m). Embryos were scored at 96 hpf (after 72 hr of pro-oxidant exposure). n ≥ 100/individual animals per group, p value from chi-square analysis. Data pooled from three independent experiments.
(C) Severe hemolytic edema in tp53M214K/M214K zebrafish exposed to 1-napthol (20 μg/5 mL). Hemoglobin staining was performed using o-dianisidine.
(D) Heatmap of RNA-seq data from untreated and 1-naphthol-treated tp53M214K/M214K zebrafish (treated from 24 to 72 hpf). Unsupervised clustering was performed on genes with log2 expression >2-fold difference. n = 5 individual animals in each group.
(E) GO term analysis using the PANTHER database and software for GO enrichment analysis for biological process indicates genes involved in metabolism are highly upregulated in tp53M214K/M214K after pro-oxidant challenge.
(F) Live image displaying erythroid precursors from gata1:DsRed × mito:GFP zebrafish at 72 hpf. Imaging performed with a Leica TCS SPE Spectral Confocal system. Scale bar represents 25 μm.
(G) Mitochondrial content of gata1:DsRed × mito:GFP and gata1:DsRed × mito:GFP × tp53M214K zebrafish. Single-cell suspensions at 72 hpf were made and mitochondrial content determined by flow cytometry measuring GFP MFI. n = 18–20 individual animals per group in two independent experiments.
(H) Lack of OXPHOS reserve in tp53M214/M214KK zebrafish. Mitochondrial OCR was measured using the Seahorse biochemical analyzer. Yellow bars indicate the amount of reserve OXPHOS, which is the difference between maximal OCR and basal OCR. Shown are the mean and SEM. n = 12–15 individual animals per group in two independent experiments. See also Figure S3D.
(I) Basal OCR is higher in tp53M214K/M214K zebrafish. Shown are the means and SD. n = 12–15 per group showing one of two independent experiments.
(J) Quantification of OXPHOS reserve indicates tp53M214K/M214K zebrafish have no reserve. Shown are the means and SD. n = 12–15 per group showing one of two independent experiments. All pro-oxidant exposure times were from 24 to 72 hpf, except (C) and (I and J), which were from 24 to 96 hpf and from 24 to 120 hpf, respectively.
(K) Pro-oxidant exposure (20 μg/5 mL) combined with oligomycin exposure leads to decreased death and edema in tp53M214K/M214K zebrafish. Embryos scored at 96 hpf after 72 hr of drug exposure. n ≥ 100/individual animals per group, p value from chi-square analysis. Data pooled from three independent experiments.
Being that TP53 controls many intracellular pathways with a wide variety of biological functions, we took an agnostic experimental approach and performed RNA-seq on tp53M214K homozygotes under oxidative stress and wild-type zebrafish to get an idea of which major cellular processes differ between them. We found 753 genes that were differentially expressed by greater than a 2-fold variance (Figure 4D, top genes shown in Table S2). We performed a gene ontology (GO) term analysis for gene enrichment and found the largest biological processes affected in tp53M214K homozygotes were “metabolic processes” (Figure 4E). Mitochondria are the central regulators of cellular metabolism and account for a majority of the cellular ATP production (McBride et al., 2006). Due to the optical clarity of the zebrafish, they offer unparalleled visualization of organelles when using fluorescent organelle-targeted reporters. Therefore, to determine if there was any dysregulation in mitochondria shape or number, we crossed Gata1:DsRed and Gata1:DsRed/tp53M214K animals to mito:GFP transgenic animals, which have a mitochondrial localization signal sequence fused to GFP, allowing for microscopic evaluation of mitochondrial size, shape, and enumeration by flow cytometry (Kim et al., 2008); the resultant offspring displayed DsRed-positive Gata1+ erythroid cells with GFP-tagged mitochondria (Figure 4F). We noticed no differences in Gata1+ erythroid cell mitochondria size or shape in tp53M214K/M214K compared with tp53+/+ animals (not shown). There was also no difference in Gata1+ erythroid cell mitochondrial content between tp53M214K/M214K and tp53+/+ animals (Figure 4G).
Although there was no effect of mutant tp53 on mitochondrial content, we hypothesized that mitochondrial function could be altered. Therefore, we interrogated total mitochondrial respiration using a Seahorse biochemical analyzer. We compared the oxidative phosphorylation (OXPHOS) metabolic profile of wild-type animals with that of tp53M214/M214K animals and found that tp53M214K/M214K mutants had a significantly elevated basal oxygen consumption rate (OCR) compared with that of wild-type (Figures 4H–4J and S3D). Due to their elevated basal OCR, the tp53M214K/M214K mutant animals were severely deficient in metabolic reserve capacity (p < 0.0001, Figure 4J). Although we can only measure OCR from the total animal and not specifically in erythroid precursors, we believe these findings translate to multiple cell types. We next utilized the ability of oligomycin, an inhibitor of H+-ATP-synthase, to reduce ATP, oxidative phosphorylation, and ROS production (Shchepina et al., 2002). We found that oligomycin treatment of pro-oxidant-exposed tp53M214K/M214K mutants led to a partial rescue in edematous phenotype (p < 0.0007, Figure 4K), highlighting the significant role that mitochondria play in ROS-mediated erythroid cell death.
These data provide a physiological explanation as to the mechanism governing the robust ROS generated and non-apoptotic death observed when tp53 is functionally mutated. That is, the mitochondria of tp53 mutants are undergoing OXPHOS at an elevated rate, and therefore lack significant reserve to handle pro-oxidant challenge above baseline. This results in overwhelming mitochondrial ROS generation and cell death.
Discussion
All living organisms that are able to perform cellular aerobic respiration undergo oxidation reactions producing oxygen-containing molecules that are highly reactive (i.e., ROS). ROS are a part of normal metabolism and are usually metabolized via enzymes such as catalase, superoxide dismutase, glutathione peroxidase, and others to protect the cell from damage due to uncontrolled ROS. Pro-oxidant exposure produces ROS beyond normal metabolism, creating an “oxidative challenge” or “stress.” Oxidative stress plays a pathological role in nearly all diseases, including cardiac, oncologic, liver, and neurological conditions, as well as the aging process (Dichi et al., 2014, Halliwell and Gutteridge, 2007, Lopez-Otin et al., 2013). The mechanisms of damage directly caused by oxidative stress include DNA breakage, covalent modification of proteins, induction of cell death pathways, and the introduction of DNA mutations (both mitochondrial and non-mitochondrial) (Pinto and Moraes, 2015).
The hematopoietic system exists in a low-oxygen environment (Morikawa and Takubo, 2015). Increases in ROS have been shown to decrease hematopoietic cell longevity and increase differentiation toward myeloid lineages. Ito et al. (2006) demonstrated that ROS, along with p38 MAPK phosphorylation, were induced in ATM−/− mice, which contributed to hematopoietic stem cell (HSC) exhaustion. They went on to show that inhibition of p38 MAPK could restore HSC function and longevity (Ito et al., 2006). Deletion of Foxo3, a regulator of antioxidant pathways, in mice allows for elevated levels of ROS and decreased numbers of lin−Sca1+Kit+, as well short- and long-term HSCs (Yalcin et al., 2008). This was accompanied by a decrease in quiescence as well as colony-forming unit ability. In agreement with our data, the authors also showed a significant increase in tp53 transcription levels associated with elevated ROS in Foxo3−/− mice.
TP53 is predominantly known as a tumor suppressor and a regulator of the cell cycle. TP53 activation occurs during “cellular stress” or DNA damage and blocks progression through the cell cycle to avoid propagating mutated DNA. If the genomic damage cannot be repaired, TP53 initiates the apoptosis pathway and avoids reproduction of abnormal cells. The role of TP53 in the management of oxidative stress and the response to oxidative stress is newly becoming appreciated. Experiments in fibroblasts from TP53-deficient mice or downregulation of TP53 with siRNA (in fibroblasts) show a significant increase in ROS, indicating an antioxidant role for TP53 (Sablina et al., 2005). Furthermore, mice lacking TP53 have increased mutation rates, elevated ROS, and develop lymphomas more readily, all of which are prevented by treatment with the antioxidant n-acetylcysteine (Sablina et al., 2005). The mechanisms of how TP53 regulates an oxidative stress response are not well understood, but both transcriptional and protein-protein control mechanisms play a role. TP53 can transactivate antioxidant genes, such as glutathione peroxidase 1 (GPX1) and sestrin2 (SESN2) (Sablina et al., 2005). Therefore, the lack of expression of key antioxidant pathways may contribute to the increase in ROS seen in TP53 deficiency.
Alternatively, our data suggest a link between TP53 and mitochondrial function. While mature erythrocytes undergo nuclear extrusion and mitophagy, erythroid precursors retain these organelles, making their study in model systems relevant (Mortensen et al., 2010, Mortensen and Simon, 2010, Zhang et al., 2011). The exact role of TP53 and mitochondrial function remains unclear. Mihara et al. (2003) showed that TP53 exerts a proapoptotic effect through interaction with Bcl-2 to promote mitochondrial permeabilization and cytochrome c release in cell lines. Loss of wild-type TP53 function has been shown to lead to elevated ROS and DNA damage, and experiments performed in cell lines without mtDNA had significantly attenuated ROS production and apoptosis, suggesting that the mitochondria were major participants in the production of ROS (Sablina et al., 2005). In contrast, Xavier et al. (2014) demonstrated that TP53 could be translocated to mitochondria and attenuate oxidative stress, mitophagy, and cytochrome c release during neuronal differentiation. While we have yet to understand the precise mechanism of action in our current model system, one of the advantages conferred from using the zebrafish model is the ability of evaluate the entire animal under pro-oxidant stress conditions, allowing all affected pathways to function and reveal the physiological outcomes of tp53 mutation. Furthermore, we believe this model is best compared with mammalian bone marrow-housed erythroid cells and not circulating mature erythrocytes due to the fact that, unlike mammalian erythrocytes, zebrafish mature red cells retain nuclei and mitochondria.
In addition, clinical data indicate a connection between tp53 and mitochondrial function. Our data are in agreement with a prior cohort study of patients with Li Fraumeni syndrome (LFS), which is caused by the germline transmission of TP53 mutation (Wang et al., 2013). In this study, both lymphocytes and myoblasts from patients with LFS demonstrated significantly higher rates of baseline mitochondrial OCR, suggesting that TP53 played a significant role in global mitochondrial function. Finally, there is also clinical evidence linking tp53 mutation to dyserythropoeisis in myelodysplastic syndrome (MDS). Kulasekararaj et al. (2013) analyzed the TP53 mutation status in over 300 patients with MDS. Within the patient characteristics analysis, they found that MDS patients with mutated TP53 were almost 15 times more apt to be transfusion dependent versus those without TP53 mutations. It is not known whether this was due to haploinsufficiency of TP53 leading to reduced cell production or an increase in erythroid cell death, but it does provide evidence for a link between TP53 mutation and aberrant erythropoiesis (Kulasekararaj et al., 2013). Furthermore, it is known that MDS patients with TP53 mutations have a higher mortality rate and poorer response to treatment than MDS patients without TP53 mutations (Jadersten et al., 2011).
In conclusion, we utilized the zebrafish as a model to demonstrate the contribution of Gata1+ erythroid cells to the oxidative stress response. We found that TP53 plays a key role in erythroid cells management of a Gata1+ pro-oxidant challenge. Mutation in tp53 allowed for basal mitochondrial respiration to occur at a significantly elevated rate, a contributing factor to the enhanced ROS and cell death we observed in tp53-mutant animals. This model allows us to observe the physiological responses to oxidative stress at the whole-organism level. Future work will focus on determining the specific mechanisms by which Tp53 can regulate mitochondrial function and how mutated tp53 dysregulates mitochondrial respiration, as well as the oxidative stress response.
Experimental Procedures
Zebrafish Husbandry
Zebrafish were raised and maintained by the University of Minnesota Zebrafish Core Facility according to standard procedures (Westerfield, 1993), with the approval of the International Animal Care and Use Committee. Wild-type fish (Segrest Farms) were bred in house, as were Tg (gata1:DsRed)sd2 (Traver et al., 2003). The gata1am651 line, also known as vlad tepes (vlt) (Lyons et al., 2002) was a gift of Dr. L. Zon. The tp53zdf1 line, also known as tp53m214k (Berghmans et al., 2005), was obtained from the Zebrafish International Resource Center.
Pro-oxidant Exposure
All zebrafish embryos were dechorionated at 24 hpf and maintained at 28°C. For gene expression experiments, groups of ten embryos were placed into six-well plates in a total volume of 5 mL of fish water. For flow cytometry experiments, individual embryos were placed into 96-well plates in a total volume of 300 μL. 1-Naphthol (Sigma-Aldrich) was dissolved in embryo water for a 250 μg/mL stock, made fresh for each experiment. Embryos were treated with 0, 10, 20, or 30 μg/5 mL fish water for the next 48–72 hr (i.e., 48–96 hpf), then were visually inspected for phenotype under a dissecting microscope, and dead embryos were noted and removed. In some experiments, pifithrin-alpha was added with 1-naphthol at 1 μM or n-acetylcysteine at 1 mM.
CellROX Assay
Embryos were euthanized in 200 mg/mL tricaine followed by placement into individual wells of a 96-well plate; 100 μL of 0.25% trypsin (Invitrogen) (pre-warmed to 37°C) was added to each well and incubated for 10 min at room temperature. Afterward, 200 μL of DMEM + 10% fetal bovine serum (FBS) was added to each well. Embryos were triturated until completely broken up. Then, 250 μL of the embryo cell suspension was transferred to a 96-well BioDot PCR plate. This plate has sufficient structural integrity to survive centrifuging. Plates were sealed with plastic adhesive covers and spun at 500 × g (2,500 rpm) for 5 min in an MPS 1000 Mini Plate Spinner centrifuge. Cell pellets were washed in 200 μL of PBS containing 2% FBS and centrifuged again for 5 min. Supernatant was removed by flicking the plate into the sink. Cells were placed in PBS containing 2% FBS with 5 μM CellROX green (Invitrogen) and incubated at room temperature for 2 hr. Cell suspension was passed through a 40 μm filter prior to flow cytometry. Quantification of CellROX green ROS probe signal was through determining the mean fluorescence intensity (MFI) (the absorption/emission maxima of CellROX green is 485/520 nm) and data expressed relative to unstressed embryos.
8-Oxo-dG Detection
Zebrafish were treated from 24 to 96 hpf with 20 μg/5 mL 1-naphthol. The genomic DNA of ten zebrafish embryos at 96 hpf was extracted using the QIAamp DNA mini kit (QIAGEN). The concentration of the genomic DNA was determined by standard absorbance at 260 nm. The amount of 8-oxo-dG present in the genomic DNA was measured using the HT 8-oxo-dG ELISA kit II from Trevigen (catalog no. 4380-096-K), and the assay procedures were based on the manufacturer's protocol. The level of 8-oxo-dG was then normalized to the amount genomic DNA used in the assay.
TUNEL Assay
For the TUNEL assay, embryos were fixed in 4% paraformaldehyde overnight at 4°C followed by three washes with PBS. Embryos were then washed in dH2O and incubated in bleaching solution (30% H2O2, 40% KOH in PBS) for 20 min at room temperature. Embryos were blocked with 3% H2O2 in methanol for 10 min at room temperature followed by PBS rinse for 5 min. Embryos were placed in “perm” solution (10 mM Tris-HCl [pH 8.0] and 10 μg/mL Proteinase K) to permeabilize for 20 min at room temperature followed by PBS rinse for 5 min. Positive controls were incubated in 100 μL of dH2O containing 200 U/mL DNase I at 37°C for 15 min followed by PBS rinse for 5 min. The Roche (Roche Applied Bioscience) in situ cell death detection kit (catalog no. 11-684-817-910C) was then used to detected apoptosis following the manufacturer’s instructions, and negative controls were labeled, but no enzyme solution was added.
o-Dianisidine Staining
Hemoglobin staining was performed as described previously (Detrich et al., 1995). Briefly, dechorionated live embryos were stained in 0.6 mg/mL o-dianisidine (Sigma-Aldrich) containing 0.01 mol/L sodium acetate (pH 4.5), 0.65% H2O2, and 40% ethanol in the dark for 15 min. Embryos were then dehydrated through graded ethanol washes of 50%, 75%, and 100% for 5 min each. Finally, embryos were cleared in benzyl benzoate and benzyl alcohol at a 2:1 ratio. Embryos were stored in the clearing solution until imaged using a Leica DMI 4000 inverted microscope with PlanAPO 1.6/0.05 NA objective (Leica Camera). Image capture was performed with Leica LAS software, and post-processing was performed using Adobe Photoshop CS6.
RNA Extraction and qRT Protocols
Total RNA was collected from homogenized 96 hpf embryos (n = 10/group, 4–6 groups per treatment condition) using Trizol reagent (Invitrogen) per the manufacturer's protocol. First-strand cDNA was synthesized from 250 ng of total RNA using SuperScript II Reverse Transcriptase (Life Technologies). qRT-PCR was performed in triplicate for tp53 using TaqMan Universal MasterMix and primers (Life Technologies), with b-actin as the endogenous control. Intron-spanning primer sets used were as follows: tp53 (Dr03112089_m1); b-actin (Dr03432610_m1).
RNA-Seq
Total RNA was isolated from embryos as described above and was quantitated with Nanodrop to reach the quantity threshold of 1 μg. Sample integrity was checked using an Agilent Bioanalyzer 2100. Each sample passing the quality control (RNA integrity number >8) was used to create a poly(A) barcoded RNA-seq library using standard protocols (Illumina TruSeq RNA Sample Preparation Kit). Several samples were pooled for each sequencing lane for Illumina Hi-Seq 2000 to generate at least 10 million paired 50-base pair reads per sample. The sequencing was performed at the University of Minnesota BioMedical Genomics Center.
RNA-Seq Analysis
Raw paired-end RNA-seq reads in fastq format were assessed for base call quality, cycle uniformity, and contamination using fastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Trimmomatic was applied to remove Illumina TruSeq sequencing adapters and reads with low phred base-calling quality (Bolger et al., 2014). Pre-processed paired-end reads were then mapped to zebrafish genome (UCSC danRer7 assembly) via Tophat 2 (Trapnell et al., 2009) using the iGenomes reference UCSC zebrafish annotation (downloaded from https://support.illumina.com/sequencing/sequencing_software/igenome.html) in GTF format. Resulting Binary Alignment and Mapping files from the mapping were analyzed via the subread:featurecount-voom/limma pipeline as described by the Sequencing Quality Control (SEQC) Consortium to quantify the expression level of each known gene in units of count and test for differential expression (SEQC/MAQC-III Consortium, 2014). A matrix of abundance levels for each gene was extracted to assess sample clustering using multidimensional scaling, which revealed sample outliers. The outliers were excluded from downstream analysis. Differential gene sets were filtered to remove those with fold changes less than 2 (up- or downregulated) and with a raw p value greater than 0.005. Differentially expressed genes were mapped to their human homologous genes using BioMart and HomoloGene (http://www.ncbi.nlm.nih.gov/homologene/) (Smedley et al., 2015). Redundant entries were manually removed. The differentially expressed human gene sets were screened for enrichment of GO terms (Gene Ontology Consortium, 2015) with IPA using curated canonical pathways from the literature.
Cell Death/Apoptosis Assay
Embryos were processed as previously to create a single-cell suspension and stained using reagents from the Annexin V Apoptosis Detection Kit APC (catalog no. 88-8007-72) following the manufacture’s protocol. Quantification of cell death and apoptosis was performed from the MFI obtained on the appropriate channel using flow cytometry.
Murine Erythroid ROS
Femurs from 8–12-week-old mice were crushed with a mortar and pestle in the presence of PBS containing 2% FBS followed by filtering through a 70 μm filter. Whole marrow cells were treated with 1 mM acrolein (Sigma) and 5 μM CellROX green for 5 hr at room temperature. Cells were next stained with FC Block (no. 14-0161-85; eBioscience) for 10 min followed by staining with Brilliant Violet 421 anti-mouse CD71 (catalog no. RI7217; Biolegend) and NucBlue (Invitrogen) for 30 min and washed once with PBS containing 2% FBS. Cells were returned to original treatment media, passed through a 40 μm filter and propidium iodide (catalog no. 537059; Calbiochem) at 1 μg/mL was added just prior to flow cytometry to discriminate dead cells. Individual samples were prepared in technical triplicates.
Seahorse Bioanalyzer
One embryo was gently added to a well in XF24 Islet Capture Microplate in 525 μL of fish water. Embryos were placed on ice for 5–10 min to reduce activity and bring them to the bottom of the wells. Using the Capture Screen Insert Tool, one capture screen was placed in each well, with the “cup” side down and the flat side up, being careful to avoid squashing the embryos. All screens were checked to ensure they were firmly in place to avoid damage to the Seahorse machine. Embryos were allowed to return to room temperature and checked for heartbeats. Compound concentrations used in OXPHOS analysis were as follows: oligomycin at 56 μM, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone at 4.5 μM, antimycin at 1 μM, and rotenone at 1 μM. Calibration/setup of the Seahorse Bioanalyzer was as follows: temperature set to 30°C, all mix times for 1 min; stand times for 10–15 min, and read times for 5 min.
Author Contributions
M.L.C. performed the majority of the experiments and wrote the first draft of the manuscript; A.K. assisted in experiment execution; Y.G., M.S., and J.W. assisted in genotyping and phenotyping animals during experiments; J.T. assisted in RT-PCR experiments; Y.Z. performed the RNA-seq analyses; and T.C.L. oversaw the project, contributed to experimental design, and performed final manuscript editing.
Acknowledgments
We gratefully thank the laboratory of Dr. Zon for sharing the vlad tepes zebrafish. Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the NIH under award number K08HL108998 (T.C.L.), ASH Junior Faculty Scholar Award (T.C.L.), the University of Minnesota Academic Health Center Seed Grant (T.C.L.), The Children's Cancer Research Fund (T.C.L.), and The Viking's Research Fund (T.C.L.).
Published: January 26, 2017
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.12.025.
Accession Numbers
The dataset is available in the GEO under accession number GEO: GSE92754.
Supplemental Information
References
- Bahary N., Zon L.I. Use of the zebrafish (Danio rerio) to define hematopoiesis. Stem Cells. 1998;16:89–98. doi: 10.1002/stem.160089. [DOI] [PubMed] [Google Scholar]
- Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Kim A.D., Violette E.P., Stachura D.L., Cisson J.L., Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov A.V., Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Liu J., Heck S., Chasis J.A., An X., Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc. Natl. Acad. Sci. USA. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culmsee C., Zhu X., Yu Q.S., Chan S.L., Camandola S., Guo Z., Greig N.H., Mattson M.P. A synthetic inhibitor of p53 protects neurons against death induced by ischemic and excitotoxic insults, and amyloid beta-peptide. J. Neurochem. 2001;77:220–228. doi: 10.1046/j.1471-4159.2001.t01-1-00220.x. [DOI] [PubMed] [Google Scholar]
- Davidson A.J., Zon L.I. The 'definitive' (and 'primitive') guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- Degterev A., Hitomi J., Germscheid M., Ch'en I.L., Korkina O., Teng X., Abbott D., Cuny G.D., Yuan C., Wagner G. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney S., Jarem D.A., Volle C.B., Yennie C.J. Chemical and biological consequences of oxidatively damaged guanine in DNA. Free Radic. Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich H.W., 3rd, Kieran M.W., Chan F.Y., Barone L.M., Yee K., Rundstadler J.A., Pratt S., Ransom D., Zon L.I. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichi I., Breganó J.W., Colado Simão A.a.N., Cecchini R. Taylor & Francis; 2014. Role of Oxidative Stress in Chronic Diseases. [Google Scholar]
- Dooley K.A., Fraenkel P.G., Langer N.B., Schmid B., Davidson A.J., Weber G., Chiang K., Foott H., Dwyer C., Wingert R.A. montalcino, A zebrafish model for variegate porphyria. Exp. Hematol. 2008;36:1132–1142. doi: 10.1016/j.exphem.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy K.T., Wickstrom E. Zebrafish tp53 knockdown extends the survival of irradiated zebrafish embryos more effectively than the p53 inhibitor pifithrin-alpha. Cancer Biol. Ther. 2007;6:675–678. doi: 10.4161/cbt.6.5.3956. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Gary R.K., Jensen D.A. The p53 inhibitor pifithrin-alpha forms a sparingly soluble derivative via intramolecular cyclization under physiological conditions. Mol. Pharmaceutics. 2005;2:462–474. doi: 10.1021/mp050055d. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M.C. Fourth Edition. Oxford University Press; 2007. Free Radicals in Biology and Medicine. [Google Scholar]
- Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jadersten M., Saft L., Smith A., Kulasekararaj A., Pomplun S., Gohring G., Hedlund A., Hast R., Schlegelberger B., Porwit A. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J. Clin. Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- Jouan-Lanhouet S., Riquet F., Duprez L., Vanden Berghe T., Takahashi N., Vandenabeele P. Necroptosis, in vivo detection in experimental disease models. Semin. Cell Dev. Biol. 2014;35:2–13. doi: 10.1016/j.semcdb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Kang K.H., Kim C.H., Choi S.Y. Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. BioTechniques. 2008;45:331–334. doi: 10.2144/000112909. [DOI] [PubMed] [Google Scholar]
- Kulasekararaj A.G., Smith A.E., Mian S.A., Mohamedali A.M., Krishnamurthy P., Lea N.C., Gaken J., Pennaneach C., Ireland R., Czepulkowski B. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br. J. Haematol. 2013;160:660–672. doi: 10.1111/bjh.12203. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons S.E., Lawson N.D., Lei L., Bennett P.E., Weinstein B.M., Liu P.P. A nonsense mutation in zebrafish gata1 causes the bloodless phenotype in vlad tepes. Proc. Natl. Acad. Sci. USA. 2002;99:5454–5459. doi: 10.1073/pnas.082695299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsee D.K., Pinkus G.S., Yu H. CD71 (transferrin receptor): an effective marker for erythroid precursors in bone marrow biopsy specimens. Am. J. Clin. Pathol. 2010;134:429–435. doi: 10.1309/AJCPCRK3MOAOJ6AT. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U.M. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Mohanty J.G., Nagababu E., Rifkind J.M. Red blood cell oxidative stress impairs oxygen delivery and induces red blood cell aging. Front. Physiol. 2014;5:84. doi: 10.3389/fphys.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa T., Takubo K. Hypoxia regulates the hematopoietic stem cell niche. Pflugers Arch. 2015 doi: 10.1007/s00424-015-1743-z. [DOI] [PubMed] [Google Scholar]
- Mortensen M., Simon A.K. Nonredundant role of Atg7 in mitochondrial clearance during erythroid development. Autophagy. 2010;6:423–425. doi: 10.4161/auto.6.3.11528. [DOI] [PubMed] [Google Scholar]
- Mortensen M., Ferguson D.J., Simon A.K. Mitochondrial clearance by autophagy in developing erythrocytes: clearly important, but just how much so? Cell Cycle. 2010;9:1901–1906. doi: 10.4161/cc.9.10.11603. [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Patrinostro X., Carter M.L., Kramer A.C., Lund T.C. A model of glucose-6-phosphate dehydrogenase deficiency in the zebrafish. Exp. Hematol. 2013;41:697–710.e2. doi: 10.1016/j.exphem.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone S., Tataranno M.L., Stazzoni G., Del Vecchio A., Buonocore G. Oxidative injury in neonatal erythrocytes. J. Maternal Fetal Neonatal Med. 2012;25:104–108. doi: 10.3109/14767058.2012.715471. [DOI] [PubMed] [Google Scholar]
- Peslak S.A., Wenger J., Bemis J.C., Kingsley P.D., Koniski A.D., McGrath K.E., Palis J. EPO-mediated expansion of late-stage erythroid progenitors in the bone marrow initiates recovery from sublethal radiation stress. Blood. 2012;120:2501–2511. doi: 10.1182/blood-2011-11-394304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M., Moraes C.T. Mechanisms linking mtDNA damage and aging. Free Radic. Biol. Med. 2015;85:250–258. doi: 10.1016/j.freeradbiomed.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S., Campbell K.J., Roche K.C., Perkins N.D. The p53-inhibitor pifithrin-alpha inhibits firefly luciferase activity in vivo and in vitro. BMC Mol. Biol. 2003;4:9. doi: 10.1186/1471-2199-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEQC/MAQC-III Consortium A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014;32:903–914. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E., DePinho R.A. p53: good cop/bad cop. Cell. 2002;110:9–12. doi: 10.1016/s0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- Shchepina L.A., Pletjushkina O.Y., Avetisyan A.V., Bakeeva L.E., Fetisova E.K., Izyumov D.S., Saprunova V.B., Vyssokikh M.Y., Chernyak B.V., Skulachev V.P. Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene. 2002;21:8149–8157. doi: 10.1038/sj.onc.1206053. [DOI] [PubMed] [Google Scholar]
- Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Humphries J.M., White R.M., Murphey R.D., Burns C.E., Zon L.I. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp. Hematol. 2012;40:228–237.e5. doi: 10.1016/j.exphem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wang P.Y., Ma W., Park J.Y., Celi F.S., Arena R., Choi J.W., Ali Q.A., Tripodi D.J., Zhuang J., Lago C.U. Increased oxidative metabolism in the Li-Fraumeni syndrome. N. Engl. J. Med. 2013;368:1027–1032. doi: 10.1056/NEJMoa1214091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. Zebrafish International Resource Center; 1993. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) [Google Scholar]
- Xavier J.M., Morgado A.L., Sola S., Rodrigues C.M. Mitochondrial translocation of p53 modulates neuronal fate by preventing differentiation-induced mitochondrial stress. Antioxid. Redox Signal. 2014;21:1009–1024. doi: 10.1089/ars.2013.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin S., Zhang X., Luciano J.P., Mungamuri S.K., Marinkovic D., Vercherat C., Sarkar A., Grisotto M., Taneja R., Ghaffari S. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- Zhang Z.W., Cheng J., Xu F., Chen Y.E., Du J.B., Yuan M., Zhu F., Xu X.C., Yuan S. Red blood cell extrudes nucleus and mitochondria against oxidative stress. IUBMB Life. 2011;63:560–565. doi: 10.1002/iub.490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.