Figure 4.
RNA-Sequencing Data Reveal Thyroid Signature
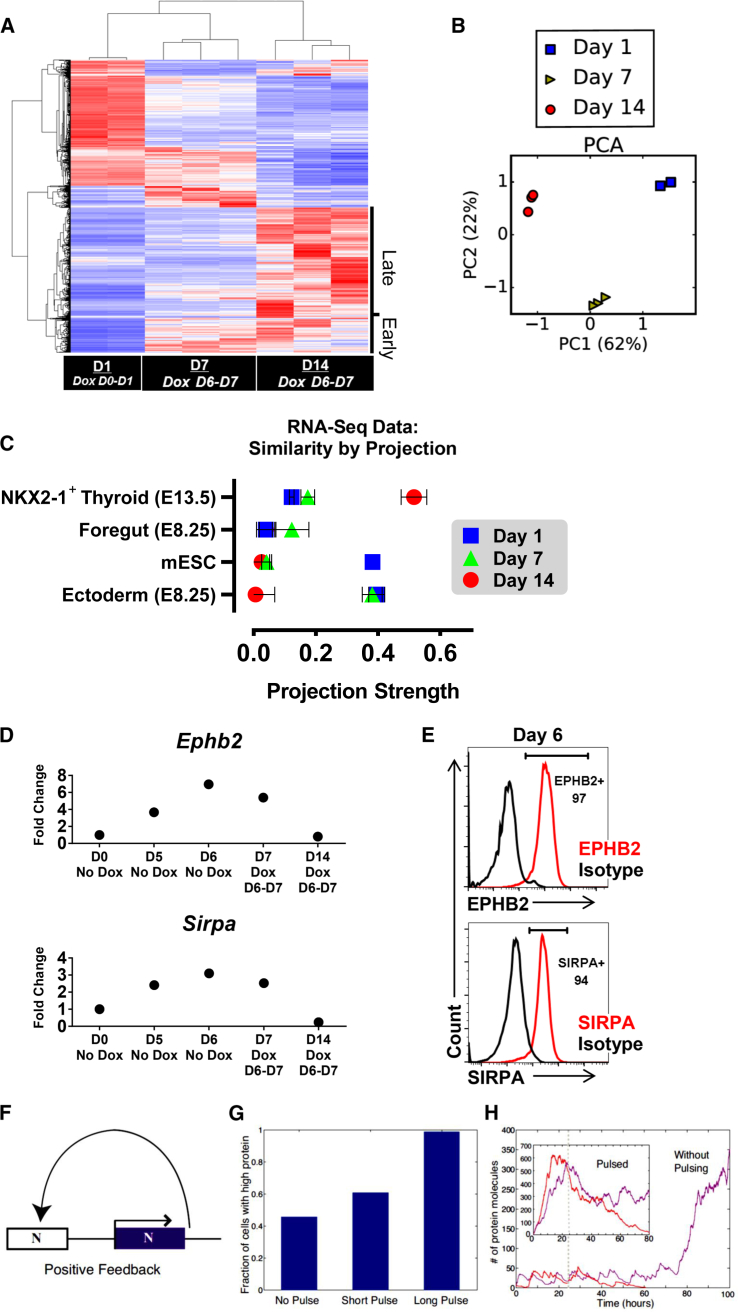
(A) RNA-seq heatmap of the top 9,088 differentially expressed genes with highest variance (false discovery rate <0.05) among samples, clustered by samples (columns) and genes (rows). Late and early clusters are indicated.
(B) PCA plot of RNA-seq populations.
(C) Projection graph representing the degree of similarity (x axis, exact match = 1) between RNA-seq samples (dots) and reference gene expression datasets (y axis).
(D) RT-qPCR validation data of cell surface markers identified in the early cluster.
(E) Flow cytometry data corresponding with (D).
(F) Bistable model: a protein that cooperatively binds its own promoter leads to a positive feedback-based, bistable switch.
(G) Percentage of high-protein-expressing cells in Monte-Carlo simulations of the bistable switch shown in (F) for three pulse lengths of protein expression. Results were calculated using 1,000 simulations in each condition.
(H) Stochastic trajectories of protein number as a function of time in the absence of pulsing (main figure) and in the presence of a long pulse (inset). Colored lines show stochastic trajectories that successfully activate the Nkx2-1 feedback loop (purple lines) or fail to activate the feedback loop (red lines). The dashed line indicates the time the basal transcription rate was turned off in all simulations.
See also Figure S4.