Figure 6. Validation of three novel ZFP36 target genes involved in EMT.
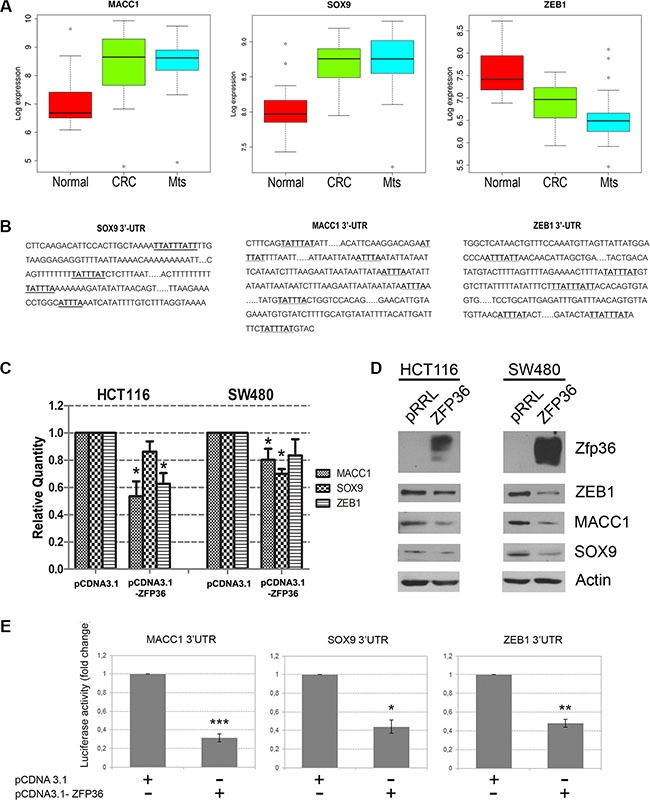
(Panel A) Boxplot of Log 2 expression values of MACC1, SOX9 and ZEB1 in 23 normal colon mucosa (Normal), 30 primary colon carcinoma (CRC) and 27 liver metastases (Mts) samples. The thick line indicates the median value, the coloured box indicates the interquartile range and the whiskers the minimum and maximum values excluded outliers. Open circles represent data points outside the whiskers. (Panel B) Schematic representation of the 3′UTRs sequences of MACC1, SOX9 and ZEB1. A-U rich sequences (ARE) are highlighted in bold. (Panel C) HCT116 and SW480 cells were transfected with an empty vector (pCDNA3.1) or a ZFP36-overexpressing vector (pCDNA3.1-ZFP36). RNA was extracted after 48 hours and MACC1, SOX9, ZEB1 mRNA levels were analysed through qRT-PCR analysis. Results are represented as means of three experiments (+/−SEM) and GAPDH was used as endogenous control. *p < 0.05. (Panel D) HCT116 and SW480 were infected with an empty vector (pRRL) or a ZFP36-overexpressing vector (ZFP36) and corresponding total protein lysates were analysed through Western blotting techniques with antibodies against ZEB1, MACC1, SOX9 and ZFP36. Actin was used as loading control. (Panel E) A fragment of the 3′UTRs of MACC1, SOX9 and ZEB1 was cloned in a pGL3 vector, downstream of the Luciferase gene. These constructs where co-transfected with a Δ-gal reporter plasmid and with an empty vector (pCDNA3.1) or ZFP36 overexpressing vector (pCDNA3.1-ZFP36) in HEK293T cells. Cells were harvested after 48 hours, luciferase activity was measured and normalized over Δ-gal signals. Results are represented as means of three independent experiments +/−SEM. *p < 0.05, **p < 0.001, ***p < 0.0001.
