Figure 1. UDG depletion causes incorporation of uracil and 5-FU into genomic DNA by 5-FdU.
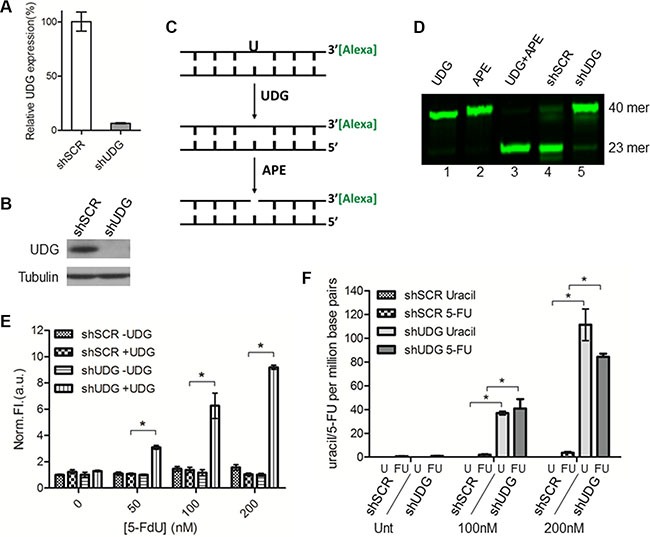
Lentiviral non-targeted scramble control shRNA (shSCR) or UDG-directed shRNA (shUDG) were transfected into DLD1 colon cancer cells, and stable cell lines were established. (A) UDG mRNA and (B) protein expression levels were determined by qPCR and western blot, respectively. The shRNA that we used targets both mitochondrial and nuclear UDG, which are collectively termed UDG in this study. (C) Schematic diagram of glycosylase activity assay by using 3′-Alexa tagged 40-mer DNA duplex with a uracil incorporation paired with adenine. (D) 10 μg nuclear extracts from DLD1 shSCR or shUDG cells were incubated with 3′-Alexa labeled oligonucleotide containing U:A base pair for 20 minutes at 37°C. Reactions with purified enzymes were used as controls. Cellular UDG activity was visualized by denaturing gel electrophoresis to separate intact 40-mer from 23-mer. (E) DLD1 shSCR and shUDG cells were treated with 0, 50, 100, and 200 nM 5-FdU for 48 h. Genomic DNA was extracted and treated in vitro with purified UDG (+ UDG) or vehicle control (− UDG). AP sites detection was performed by incubation of DNA with a cyanine-based AP site probe. Data represent mean and SD of relative fluorescence intensity normalized to 5-FdU untreated shSCR -UDG sample from three independent experiments. (*P < 0.05) (F) DLD1 shSCR and shUDG cells were untreated (Unt) or treated with 5-FdU 100 and 200 nM for 48 h. Genomic DNA was extracted and incubated in vitro with purified UDG enzyme. Uracil and 5-FU were quantified by LC-MS/MS as described in the Materials and Methods. Data represent mean and SD from three independent experiments. (*P < 0.05).
