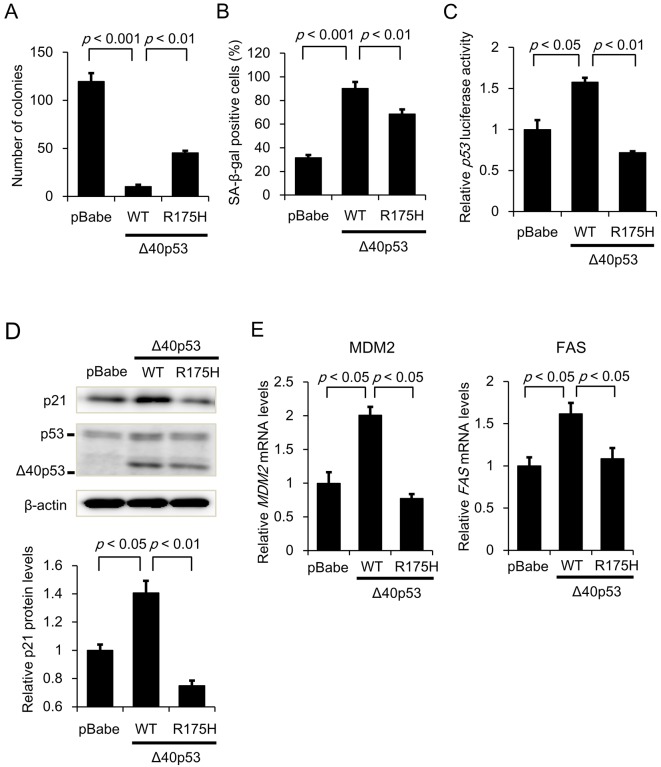
Fig. 4.
Involvement of transcriptional activity in the tumor suppressor activity of Δ40p53. (A) Colony formation assay. pBabe, Δ40p53 (WT)/pBabe, and Δ40p53 (R175H)/pBabe retroviruses were generated using 293T cells. After the viral supernatants were prepared, HepG2 cells were infected as described in the legend of Fig. 3B. Bar graphs represent the number of stained colonies (mean±s.e.m.; n=6). (B) SA-β-gal assay. pBabe/HepG2, Δ40p53 (WT)/HepG2, and Δ40p53 (R175H)/HepG2 cells (2×104 cells/well) were seeded in 12-well plates. The cells were incubated for 48 h and then stained. Bar graphs represent the percentage of SA-β-gal-positive cells (mean±s.e.m.; n=3). (C) p53-dependent transactivation was examined using a p53 luciferase reporter assay. Each clone was co-transfected with 0.15 μg of the TG13-Luc vector (containing the WT p53 DNA-binding site; p53 firefly activity) and 0.03 μg of the phRL-TK vector (internal control; Renilla luciferase activity). Luciferase activity was measured 48 h after the transfection. After normalization to Renilla luciferase activity, the data are expressed relative to the p53-dependent luciferase activity in the HepG2/RI #1 cells, which was arbitrarily defined as 1 (mean±s.e.m.; n=4). (D) Protein levels of p21 and p53 isoforms were examined by western blot analysis as described in Fig. 1F. β-actin was used as an internal control. (E) qRT-PCR analysis of MDM2 and FAS gene expression. The relative gene expression levels are shown after normalization to GAPDH mRNA expression. Data are presented relative to the mRNA expression in pBabe/HepG2 cells, which was arbitrarily defined as 1 (mean±s.e.m.; n=3).