Abstract
Recent findings from solid state NMR (ssNMR) studies suggested that the (R)-enantiomer of rimantadine binds to the full M2 protein with higher affinity than the (S)-enantiomer. Intrigued by these findings, we applied functional assays, such as antiviral assay and electrophysiology (EP), to evaluate the binding affinity of rimantadine enantiomers to the M2 protein channel. Unexpectedly, no significant difference was found between the two enantiomers. Our experimental data based on the full M2 protein function were further supported by alchemical free energy calculations and isothermal titration calorimetry (ITC) allowing an evaluation of the binding affinity of rimantadine enantiomers to the M2TM pore. Both enantiomers have similar channel blockage, affinity, and antiviral potency.
Keywords: Rimantadine enantiomers, isothermal titration calorimetry, free energy perturbation, Bennett’s acceptance ratio, electrophysiology, synthesis, antiviral assay, membrane protein, influenza M2 pore
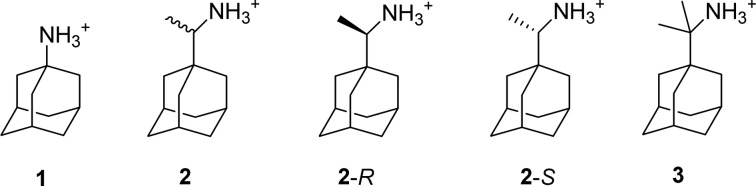
Amantadine (1) and rimantadine (2) (Scheme 1) are channel blockers of proton transit by the influenza virus M2 proton channel1,2 and long used prophylactics and therapeutics against influenza A viruses.3 The primary binding site of 1 and 2 is the lumen of the transmembrane domain of a tetrameric M2 protein (M2TM: amino acids 22–46) that forms the proton transit path.4
Scheme 1. Structures of Studied Aminoadamantane Derivatives Amantadine (1), Rimantadine (2, 2-R, and 2-S), and Rimantadine Analogue 3.
Although 1 and 2 have been used as antivirals for decades, it was only after 2008 that high resolution structures from X-ray and ssNMR experiments unveiled the structures of M2TM in complex with 1 or 2.5−9 According to these findings, the M2TM protein channel is blocked by 1 or 2 via a pore-binding mechanism.6−10 The adamantane cage in 1 or 2, as well as in other aminoadamantane analogues,11−13 is tightly contacted on all sides by V27 and A30 side chains, producing a steric occlusion of proton transit6−9 and thereby preventing the viral replication. The ssNMR results for 2 also demonstrated that the ammonium group of the drug is pointing toward the four H37 residues at the C-terminus.9 This orientation can be stabilized either through hydrogen bonds between the ammonium group of the aminoadamantane ligand and water molecules in the channel lumen which exist between the imidazoles of H37 and the ligand,13 and/or with A30 carbonyls in the vicinity,14 according to experimental9,14−16 and MD simulations data.13,17−21 Provided that M2TM is a minimal model for M2 binding,10 these high resolution structures can be used for the development of new ligands which may bind more effectively to the M2TM pore.
The effect of ligand’s chirality in its binding with a chiral receptor is of outstanding significance and the characterization of protein–ligand interactions for each enantiomer separately may identify potential stereospecific binding interactions to the receptor. While rimantadine analogues are known antiviral drugs for more than four decades, the relative potency of rimantadine enantiomers has not been studied at the molecular level. The binding affinity of each enantiomer results from chiral interactions with the binding area inside the 4-fold symmetric M2 protein. Based on differences in isotropic chemical shift changes measured using ssNMR and MD simulations results, it has been recently suggested that 2-R and 2-S have a strong but differential binding to full length M2, i.e., that 2-R binds more tightly than 2-S.22 This was the first state of the art ssNMR study of the full M2 protein and analysis of the rimantadine enantiomers binding by ssNMR, but this conclusion appears to be puzzling because: (1) 2-R and 2-S have similar in vivo antiviral activity in protecting mice from lethal influenza;23 (2) rimantadine was developed prior to the 1992 FDA guidance on the development of stereoisomers. It was approved as commercial drug in the US in 1993 containing both enantiomers. To solve the controversy between the in vitro binding assay, i.e., ssNMR results, and the in vivo efficacy results, we hereby characterize the binding affinity of the two rimantadine enantiomers and their antiviral efficacy using a consortium of in vitro and cellular assays and biophysical/computational studies.
Taken together, our results demonstrate that rimantadine enantiomers (2-R and 2-S) have equal potency. Since the controversy aroused from previous ssNMR studies of 2-R, 2-S bound to the full M2 protein, we first provided data including the antiviral activity of 2-R, 2-S against amantadine-sensitive influenza A virus strains and the blocking effect of the enantiomers against M2 using EP. Then we investigated the effect of the enantiomers to M2TM using biophysical techniques. More specifically, the antiviral potency of compounds 2-R, 2-S, and 3 against amantadine-sensitive A/Udorn/72 and A/WSN/33-M2-N31S viruses was measured. The blocking effect of 2-R, 2-S, and 3 against full length A/Udorn/72 M2 protein (M2Udorn/72) and A/WSN/33-M2-N31S protein (M2WSN/33-N31S) conductance was measured using EP via a two-electrode voltage clamp (TEVC) assay. Compound 3 is a nonchiral dimethyl analogue of rimantadine, and it was designed and synthesized as a probe to independently validate the biophysical assay results from 2-R and 2-S, as described below. The ITC binding affinities of 2-R, 2-S, and 3 against the M2TMUdorn/72 in its closed form at pH 8 were measured. Following up, we applied a free energy perturbation coupled with MD simulations (FEP/MD) scheme to calculate the relative free energies of binding between the rimantadine enantiomers (2-R and 2-S) as well as compound 3 to M2TMUdorn/72. In particular, we calculated the relative free energies of binding for the alchemical transformations of 3 to 2-R and 3 to 2-S.
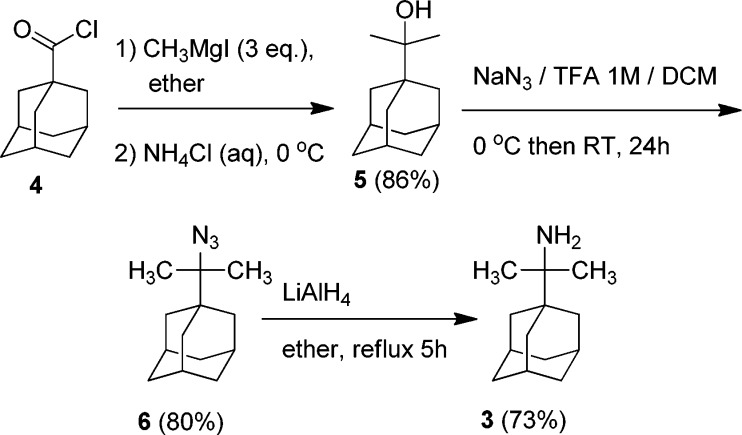
For the synthesis of primary tert-alkyl amine 3 (AdCMe2NH2), the tert-alkyl alcohol 5 was prepared according to Scheme 2 from the Grignard reaction between 1-adamantane carbonyl chloride 4 and methylmagnesium iodide. Treatment of tert-alkyl alcohol 5 with NaN3/CF3CO2H in dichloromethane afforded azide 6 in high yield, which was further subjected to reduction through LiAlH4 to form tert-alkyl amine 3 in a good yield.
Scheme 2. Synthetic Scheme for the Preparation of tert-Alkyl Amine 3.
An antiviral assay was used24,25 to compare the antiviral activity of 1, 2, 2-R, 2-S, and 3 against two amantadine-sensitive influenza A strains, A/Udorn/72 (H3N2), and A/WSN/33-M2-N31S26 (H1N1) in MDCK cells. All compounds showed submicromolar EC50 values against both influenza strains, and there was no significant difference between the two rimantadine enantiomers 2-R and 2-S (Table 1).
Table 1. Antiviral Activity of Compounds 1–3 against Influenza Virus A/Udorn/72 (H3N2) and A/WSN/33-M2-N31S (H1N1) in Madin–Darby Canine Kidney Cells.
| compd | IC50 (μM)a (A/Udorn/72) | IC50 (μM)a (A/WSN/33-M2-N31S) |
|---|---|---|
| 1 | 0.33 ± 0.04 | 0.48 ± 0.05 |
| 2 | 0.05 ± 0.02 | 0.04 ± 0.02 |
| 2-R | 0.05 ± 0.01 | 0.04 ± 0.01 |
| 2-S | 0.06 ± 0.02 | 0.02 ± 0.01 |
| 3 | 0.04 ± 0.01 | 0.03 ± 0.02 |
Mean and standard deviations of the 50% inhibitory concentration (IC50) of at least three independent measures.
The inhibitors were tested via TEVC assay using X. laevis frog oocytes microinjected with RNA expressing the M2 protein as in a previous report.27 Specifically, the blocking effect of the aminoadamantane derivatives against M2 was investigated with electrophysiology experiments using M2Udorn/72 (Table 2). The potency of the inhibitors was expressed as the inhibition percentage of the A/M2 current observed after 2 and 5 min of incubation with 100 μM compound. The electrophysiology experiments against M2Udorn/72 showed that 2-R and 2-S block the M2 channel equally to amantadine (about 90%) at a concentration of 100 μM. In addition, we also tested these aminoadamantane analogues in inhibiting another amantadine-sensitive M2 channel, M2WSN/33-N31S, which contains the V28I mutation in M2TM sequence, with an electrophysiology assay. Again, no significant difference in channel blockage after 5 min was found among 2-R, 2-S, and 3. As discussed previously,27 these measurements at 2 or 5 min are made prior to the establishment of equilibrium due to very slow on and off rates for entry of thedrugs into the constricted M2 channel and the problems of maintaining cells at low pH for extended periods.
Table 2. Block of Full-Length M2-Dependent Current by Adamantane Analoguesa.
| M2Udorn/72 |
M2WSN/33-N31S |
|||
|---|---|---|---|---|
| compd | % block after 2 min | % block after 5 min | % block after 2 min | % block after 5 min |
| 1 | 90 ± 2% (100 μM; 3) | 95 ± 1% (100 μM; 3) | ||
| 2 | 96 ± 1% (100 μM; 3) | 96 ± 1% (100 μM; 3) | 84 ± 1% (100 μM; 3) | 93 ± 0% (100 μM; 3) |
| 2-R | 93 ± 1% (100 μM; 3) | 95 ± 1% (100 μM; 3) | 71 ± 1% (100 μM; 3) | 90 ± 1% (100 μM; 3) |
| 2-S | 95 ± 1% (100 μM; 3) | 96 ± 1% (100 μM; 3) | 78 ± 1% (100 μM; 4) | 92 ± 0% (100 μM; 4) |
| 3 | 90 ± 2% (100 μM; 3) | 96 ± 1% (100 μM; 4) | 56 ± 3% (100 μM; 3) | 80 ± 2% (100 μM; 3) |
For each compound, percent block of pH-dependent M2 current at listed concentrations (± SEM). Parentheses show number of replicates.
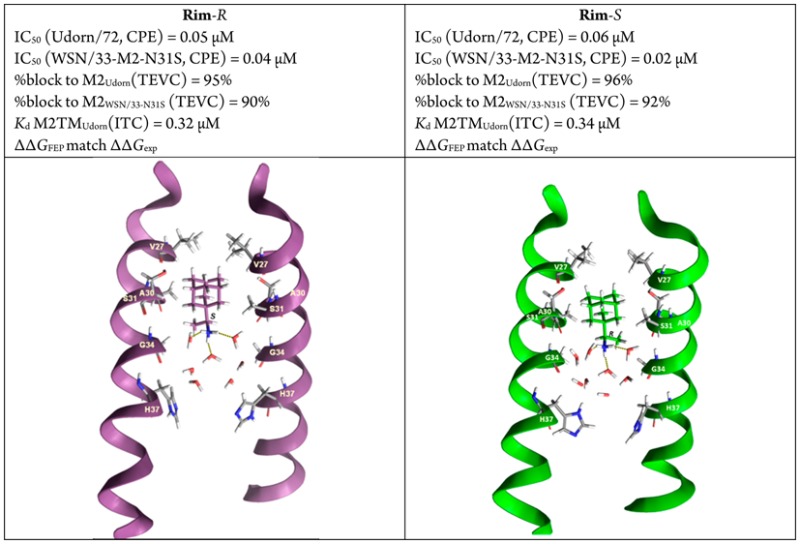
Table 3 includes thermodynamic parameters of binding against M2TMUdorn/72. Binding affinities were determined by ITC28 for M2TM-ligand systems in dodecylphosphocholine (DPC) micelles at pH 8, where M2TM fragments form stable tetramers.29 ITC measurements yield the enthalpy of binding (ΔH) as well as the dissociation constant (Kd). From Kd, the binding free energy (ΔG) is calculated (Table 3). The estimation of the binding entropy is based on the difference between ΔG and ΔH. Binding constants of 1, 2-R, 2-S, and racemate 2 were measured in a previous work13 and were included in Table 1. The enantiomers 2-R and 2-S may have a different enthalpy of binding against M2TM protein since the two complexes formed are diastereomers.30 As depicted in Table 3, enantiomers 2-R and 2-S have the same Kd values against M2TMUdorn/72 (Kd = 0.34 and 0.32 μM respectively). These values are close to the Kd = 0.51 μM of the racemic 2,13 considering the errors of the measurements and that the commercially available enantiomers have a lower chemical purity compared to racemic 2 (see Table 3, note g). An effect from the impurities on the ITC results of 2-R and 2-S cannot be excluded. However, we do not expect large changes in the measured Kd values for 2-R and 2-S, given also the very similar/identical affinity results of the two enantiomers from TEVC and antiviral assays. Compound 3, having two methyl groups instead of one methyl group in 2, has the smallest Kd (0.13 μM), i.e., the highest binding affinity of all studied aminoadamantane compounds, suggesting that polar and lipophilic characteristics are well balanced in its structure. All three compounds (2-R, 2-S, and 3) were more potent than amantadine (1).
Table 3. Binding Constant, Free Energy, Enthalpy, and Entropy of Binding Derived from ITC Measurements for M2TMUdorn/72.
| liganda | Kdb | ΔGc,d | ΔHc,e | –TΔSc,f |
|---|---|---|---|---|
| 1 | 2.17 ± 0.52 | –7.77 ± 0.14 | –6.66 ± 0.50 | –1.11 ± 0.52 |
| 2 | 0.51 ± 0.26 | –8.64 ± 0.30 | –7.60 ± 0.28 | –1.04 ± 0.41 |
| 2-Rg | 0.32 ± 0.16 | –8.97 ± 0.26 | –7.54 ± 0.34 | –1.42 ± 0.43 |
| 2-Sg | 0.34 ± 0.12 | –8.88 ± 0.21 | –7.73 ± 0.28 | –1.15 ± 0.35 |
| 3 | 0.13 ± 0.12 | –9.30 ± 0.43 | –4.19 ± 0.28 | –5.12 ± 0.51 |
Binding constant Kd in μM calculated from measured Ka in M–1 by Kd = 1/Ka × 10–6 and error in Kd in μM determined by Kd, error = (Ka, error/Ka2) × 10–6 (ITC measurements were performed in triplicate for each ligand to calculate means and standard deviations).
In kcal mol–1.
Free energy of binding
computed
from Kd by ΔG =
−RT ln(Kdref/Kd) with Kdref = 1 M and T = 300 K and
error in ΔG determined according to 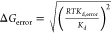 with T = 300 K.
with T = 300 K.
Enthalpy of binding and error in the enthalpy of binding calculated from measured binding enthalpy and measured error by ΔH = ΔHmeasured (T/Tmeasured) with T = 300 K and the temperature at which the ITC measurements were performed Tmeasured = 293.15 K.
Entropy of
binding calculated by
ΔS = (−ΔG + ΔH)/T and error in ΔS computed by the equation 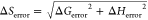 .
.
The purity of each enantiomer used was 90% for 2-R and 95% for 2-S; the enantiomeric excess (ee) of both 2-R and 2-S is 99% (Mosher’s method); the purity of compound 3 used was >99%.
We then analyzed the binding properties of 2-R and 2-S by performing alchemical free energy calculations13,31 using the Bennett acceptance ratio (BAR) method.32,33 The calculations for the alchemical transformations 3 → 2-R, 3 → 2-S were run, and the results of the FEP/MD predictions were compared with binding affinities measured by ITC (Table 4). The computational predictions were set by employing a protocol successfully benchmarked by our group in order to match experimental conditions as closely as possible.13 M2TMUdorn/72 structure was simulated in the closed conformation found at high pH and after assigning a neutral form for all H37. M2TMUdorn/72-ligand complexes were simulated in DMPC bilayers, which represent an optimal membrane mimetic system for retaining proper M2TM structure compared to other glycerophospholipids.34 We used an experimental structure (PDB ID 2KQT(5,7)) determined at pH 7.5 in DMPC bilayers; the restraints of the apo-M2TM structure were originally measured by the Cross group.5 This M2TMUdorn/72 structure is already adapted to these environmental conditions, and thus only a short equilibration phase is required. The experimental relative binding free energy values (ΔΔGexp) for the transformations 3 → 2-R and 3 → 2-S were 0.33 and 0.42 kcal mol–1 (Table 4), favoring 3. The experimental relative binding free energy values were quite close to calculated values (ΔΔGFEP) of 0.62 and 0.68 kcal mol–1, respectively (the accuracy of the calculations method is ∼1 kcal mol).32,33 The calculations also predict the experimental finding by ITC that 2-R and 2-S have the same binding affinity against M2TMUdorn/72 (Table 3), which is consistent with the results from aforementioned antiviral and electrophysiology assays performed using full M2 protein (Tables 1 and 2). Thus, using the functional core of M2 (transmembrane domain) for binding studies is appropriate.
Table 4. Relative Binding Free Energies for Pairs of Compounds Computed with the BAR Method for M2TM Embedded in a DMPC Bilayer or Derived from Experimental Binding Affinity Data in Table 3.
| M2TMUdorn/72 |
||
|---|---|---|
| transformation | ΔΔGFEPa,b | ΔΔGexpa,c |
| 3 → 2-R | 0.62 ± 0.14 | 0.33 ± 0.50 |
| 3 → 2-S | 0.68 ± 0.15 | 0.42 ± 0.48 |
In kcal mol–1.
Propagation error calculated according to the bootstrap method.31
Difference in binding free energy
calculated from experimentally determined Kd values by ΔΔG = −RT ln (KdA/KdB) with T = 300 K and error
calculated from individual experimental errors by 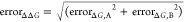 .
.
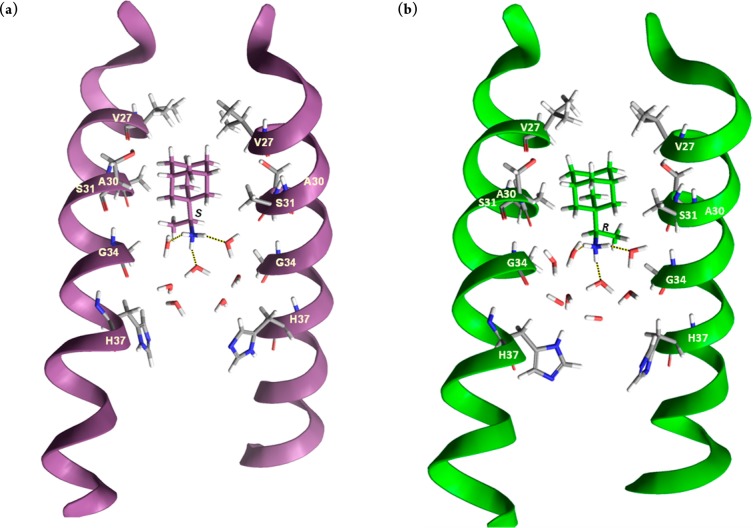
The simulated complexes of M2TMUdorn/72 with 2-R, 2-S, and 3 showed that the height of the ligands inside the pore differed only slightly, that is less than 0.3 Å toward the N-terminus relatively to 1 (Table S1), and the orientation of 2-R and 2-S in the pore differs only slightly, in accordance to their induced similar ssNMR chemical shifts for S31 and G34 when complexed with M2.22 The center of mass between the four V27 and the adamantyl ring of the ligand (V27-Ad) varies between 4.0 and 4.5 Å on average (Table S1). For 2-R and 2-S the average tilt angle was measured ∼14°, in accordance to experimental ssNMR values,9 and for 3 the average tilt angle was measured ∼5°. The angle between the pore axis or the normal of the membrane and C–N bond vector was ∼11° for 1 and close to 50° for 2-R, 2-S, and 3. These angle values suggest that the ammonium group of all aminoadamantane compounds is oriented toward the C-terminus, in consistency with previous experimental findings6,7,9 and MD simulations13,31,35 (Table S1). The distance between the adamantyl ring and the center of mass between the four A30 (Ad-A30) for 1–3 was measured ∼1 Å, and the distance CH3(lig.)-G34Ca for 2-R, 2-S, and 3 was 2.9, 3.2 and 2 Å respectively, close to the REDOR measurements for 2-R, 2-S.22 The adamantyl ring was embraced by the V27 and A30 side chains, which defined the binding site of the ligands. Compounds 1, 2-R, 2-S, and 3 form hydrogen bonds (average of three hydrogen bonds) through the ammonium group with neighboring water molecules which are positioned between the ligand and H37 residues. In the area located below the adamantyl ring toward the N-terminus no waters were found,13,21,31,35 which is consistent with the proton blocking effect of 1 and other aminoadamantane derivatives.1,12,27 A snapshot from the simulation of ligands 2-R and 2-S is depicted in Figure 1. The CHCH3 fragment, which includes the chiral carbon of 2-R, 2-S, fits into the cleft between G34 and A30, with the later being a chiral amino acid that can differentiate binding interactions between the two enantiomers. However, the distance CH3(lig.)-A30CH3, which is similar for both enantiomers (3.9 ± 0.3 and 3.5 ± 0.3 Å), suggests no difference in their van der Waals interactions. The measures suggest that hydrogen bonding interactions for 2-R, 2-S and geometric measures, which reflect van der Waals contacts, were found to be similar for both enantiomers (see Table S1). These measures are consistent with the calculated relative binding affinities, which are in accordance to the ITC data and functional assays described previously.
Figure 1.
Representative replicas from the simulation (a) of 2-S and (b) of 2-R bound to M2TMUdorn/72. Seven and eight waters are shown between the ligand and H37 residues, respectively. Three hydrogen bonds between the ammonium group of the ligand and three water molecules are shown. Hydrogen bonding together with van der Waals interactions of the adamantane core with V27 and A30 stabilize the ligand inside the pore with its ammonium group oriented toward the C-terminus.
Our results demonstrated no difference in the binding affinity between the two enantiomers, whereas the recent ssNMR study22 concluded that 2-R is a stronger binder than 2-S. According to our best understanding, the authors in ref (22) made the following important observations in the ssNMR spectra regarding the relative binding of 2-R and 2-S. In the presence of 2-R or 2-S a new resonance (i.e., one that was not recorded in the spectrum of the apo-M2 protein) was observed for S31 at 120/64 ppm in the 15N/13Ca spectrum, which had strong intensity for both enantiomers. The G34 resonance had also a stronger intensity for both enantiomers compared to the spectrum of the apo-M2. The S31 and G34 resonance frequencies were similar for both 2-R and 2-S. These results provided an experimental evidence that both enantiomers bind strongly and the binding site and orientation of the drug in the pore are similar for the two enantiomers as mentioned in ref (22). However, the authors reported the appearance of an additional resonance of medium intensity for the 2-S enantiomer at 115/63 ppm close to the frequencies of the S31 resonance of the unbound M2 state at ∼114/62 ppm and suggested a weaker binding of 2-S. Possibly the results published by Wright et al.22 are not in full agreement with those reported here as a consequent of the different methodology they applied, i.e., ssNMR spectroscopy.
In conclusion, our results showed that rimantadine enantiomers (2-R and 2-S) bind equally well to the M2 proton channel and have equal channel blockage and antiviral activity against amantadine-sensitive M2 channels. This conclusion was supported by a consortium of techniques including antiviral assays, electrophysiology ITC, and FEP/MD. Our nonclinical results support the previous use of rimantadine as a racemic mixture drug for the prevention and treatment of influenza virus infection. Further correlation of these results with the pharmacokinetic and pharmacodynamic properties for each enantiomer in humans would confirm these findings. For example, although there are no significant differences in the concentration–time profiles and disposition of 2-R and 2-S and of the 3-hydroxyrimantadine metabolites,38 large stereospecific differences in the disposition of their 4-hydroxyrimantadine metabolites are observed.36 However, it should be noted that 3- and 4-hydroxy metabolites, both of which are found in rimantadine-treated patients, showed only modest inhibitory activity against influenza A virus, i.e., they are modestly active metabolites.37
Acknowledgments
We are grateful to Chiesi Hellas for supporting this research and the Ph.D. work of C.T.; J.W. thanks the support from NIH AI119187 and the PhRMA Foundation 2015 Research Starter Grant in Pharmacology and Toxicology; A.H. and M.S. thank Andreas Sauerbrei for continuous kind support.
Glossary
Abbreviations
- M2TM
residues 22–46 of M2 protein comprising the transmembrane domain
- CPE assay
cytopathic effect assay
- ssNMR spectroscopy
solid state Nuclear Magnetic Resonance spectroscopy
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- FEP
free energy perturbation
- MD
molecular dynamics
- REDOR experiment
rotational echo double resonance experiment
- ITC
isothermal titration calorimetry
- EP
electrophysiology
- TEVC assay
two-electrode voltage clamp assay
- BAR
Bennett acceptance ratio
- PDB
protein data bank
- IC50
50% inhibitory concentration
- ee
enantiomeric excess
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.6b00311.
Supplementary material including computational and experimental protocols, as well as one table with measures from simulations, and references (PDF)
Author Present Address
(A.D.) Pharmaceutical and Medicinal Chemistry, Institute of Pharmacy and Food Chemistry, Julius-Maximilians-Universität Würzburg, Am Hubland, 97074 Würzburg, Germany.
Author Contributions
A.D. and C.T. contributed equally. This research includes part of the Master thesis of A.D. and part of the Ph.D. work of C.T. A.K. designed this research project. A.D. and C.T. both did FEP calculations and ligand synthesis. C.M. and Y.H. in J.W. group did the detailed EP experiments against the A/Udorn/72 and A/WSN/33-M2-N31S M2 and CPE assay testing against A/Udorn/72 virus. K.F. in G.G. group did the ITC measurements. A.H. in M.S. group performed CPE inhibitory assays with A/WSN/33-M2-N31S virus. A.K. wrote the manuscript, and J.W., M.S., A.H., and A.D. revised it.
The authors declare no competing financial interest.
Supplementary Material
References
- Wang C.; Takeuchi K.; Pinto L. H.; Lamb R. A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J. Virol. 1993, 67, 5585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhmakov I. V.; Geraghty F. M.; Ogden D. C.; Hayhurst A.; Antoniou M.; Hay A. J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996, 494, 329–36. 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F. G. Clinical applications of antiviral agents for chemoprophylaxis and therapy of respiratory viral infections. Antiviral Res. 1985, 5 (Suppl 1), 229–39. 10.1016/S0166-3542(85)80033-4. [DOI] [PubMed] [Google Scholar]
- Wang J.; Qiu J. X.; Soto C.; DeGrado W. F. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr. Opin. Struct. Biol. 2011, 21, 68–80. 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.; Asbury T.; Achuthan S.; Li C.; Bertram R.; Quine J. R.; Fu R.; Cross T. A. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys. J. 2007, 92, 4335–43. 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer A. L.; Acharya R.; Salom D.; Levine A. S.; Di Costanzo L.; Soto C. S.; Tereshko V.; Nanda V.; Stayrook S.; DeGrado W. F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451, 596–9. 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Schmidt-Rohr K.; Wang J.; Soto C. S.; Degrado W. F.; Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689–92. 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielak R. M.; Oxenoid K.; Chou J. J. Structural investigation of rimantadine inhibition of the AM2-BM2 chimera channel of influenza viruses. Structure 2011, 19, 1655–1663. 10.1016/j.str.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D.; Wang J.; Wu Y.; DeGrado W. F.; Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J. Am. Chem. Soc. 2011, 133, 4274–84. 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Polishchuk A. L.; Ohigashi Y.; Stouffer A. L.; Schön A.; Magavern E.; Jing X.; Lear J. D.; Freire E.; Lamb R. A.; DeGrado W. F.; Pinto L. H. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 12283–12288. 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolocouris N.; Foscolos G. B.; Kolocouris A.; Marakos P.; Pouli N.; Fytas G.; Ikeda S.; De Clercq E. Synthesis and antiviral activity evaluation of some aminoadamantane derivatives. J. Med. Chem. 1994, 37, 2896–2902. 10.1021/jm00044a010. [DOI] [PubMed] [Google Scholar]
- Kolocouris A.; Tzitzoglaki C.; Johnson F. B.; Zell R.; Wright A. K.; Cross T. A.; Tietjen I.; Fedida D.; Busath D. D. Aminoadamantanes with persistent in vitro efficacy against H1N1 (2009) influenza A. J. Med. Chem. 2014, 57, 4629–39. 10.1021/jm500598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis H.; Drakopoulos A.; Tzitzoglaki C.; Homeyer N.; Kolarov F.; Gkeka P.; Freudenberger K.; Liolios C.; Gauglitz G.; Cournia Z.; Gohlke H.; Kolocouris A. Alchemical free energy calculations and isothermal titration calorimetry measurements of aminoadamantanes bound to the closed state of influenza A/M2TM. J. Chem. Inf. Model. 2016, 56, 862–876. 10.1021/acs.jcim.6b00079. [DOI] [PubMed] [Google Scholar]
- Andreas L. B.; Barnes A. B.; Corzilius B.; Chou J. J.; Miller E. A.; Caporini M.; Rosay M.; Griffin R. G. Dynamic nuclear polarization study of inhibitor binding to the M2(18–60) proton transporter from influenza A. Biochemistry 2013, 52, 2774–82. 10.1021/bi400150x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M.; Cross T. A.; Zhou H.-X. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13311–13316. 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W.; Hong M. Conformational changes of an ion channel detected through water-protein interactions using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010, 132, 2378–84. 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intharathep P.; Laohpongspaisan C.; Rungrotmongkol T.; Loisruangsin A.; Malaisree M.; Decha P.; Aruksakunwong O.; Chuenpennit K.; Kaiyawet N.; Sompornpisut P.; Pianwanit S.; Hannongbua S. How amantadine and rimantadine inhibit proton transport in the M2 protein channel. J. Mol. Graphics Modell. 2008, 27, 342–348. 10.1016/j.jmgm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Khurana E.; Devane R. H.; Dal Peraro M.; Klein M. L. Computational study of drug binding to the membrane-bound tetrameric M2 peptide bundle from influenza A virus. Biochim. Biophys. Acta, Biomembr. 2011, 1808, 530–537. 10.1016/j.bbamem.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonov H.; Astrahan P.; Krugliak M.; Arkin I. T. How Do aminoadamantanes block the influenza M2 channel, and how does resistance develop?. J. Am. Chem. Soc. 2011, 133, 9903–9911. 10.1021/ja202288m. [DOI] [PubMed] [Google Scholar]
- Gianti E.; Carnevale V.; DeGrado W. F.; Klein M. L.; Fiorin G. Hydrogen-bonded water molecules in the M2 channel of the influenza a virus guide the binding preferences of ammonium-based inhibitors. J. Phys. Chem. B 2015, 119, 1173–83. 10.1021/jp506807y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Ma C.; Fiorin G.; Carnevale V.; Wang T.; Hu F.; Lamb R. A.; Pinto L. H.; Hong M.; Klein M. L.; DeGrado W. F. Molecular dynamics simulation directed rational design of inhibitors targeting drug-resistant mutants of influenza A virus M2. J. Am. Chem. Soc. 2011, 133, 12834–12841. 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleed M. L.; Ioannidis H.; Kolocouris A.; Busath D. D. Resistance-mutation (N31) effects on drug orientation and channel hydration in amantadine-bound influenza A M2. J. Phys. Chem. B 2015, 119, 11548–59. 10.1021/acs.jpcb.5b05808. [DOI] [PubMed] [Google Scholar]
- Wright A. K.; Batsomboon P.; Dai J.; Hung I.; Zhou H.-X.; Dudley G. B.; Cross T. A. Differential binding of rimantadine enantiomers to influenza A M2 proton channel. J. Am. Chem. Soc. 2016, 138, 1506–1509. 10.1021/jacs.5b13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich P. E.; Hermann E. C.; Meier W. E.; Paulshock M.; Prichard W. W.; Snyder J. A.; Watts J. C. Antiviral agents. 2. Structure-activity relationships of compounds related to 1-adamantanamine. J. Med. Chem. 1971, 14, 535–543. 10.1021/jm00288a019. [DOI] [PubMed] [Google Scholar]
- Torres E.; Duque M. D.; Vanderlinden E.; Ma C.; Pinto L. H.; Camps P.; Froeyen M.; Vázquez S.; Naesens L. Role of the viral hemagglutinin in the anti-influenza virus activity of newly synthesized polycyclic amine compounds. Antiviral Res. 2013, 99, 281. 10.1016/j.antiviral.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke M.; Schnittler U.; Jahn B.; Dahse H.-M.; Stelzner A. A rapid assay for evaluation of antiviral activity against coxsackie virus B3, influenza virus A, and herpes simplex virus type 1. J. Virol. Methods 2001, 95, 133–143. 10.1016/S0166-0934(01)00305-6. [DOI] [PubMed] [Google Scholar]
- Schade D.; Kotthaus J.; Riebling L.; Kotthaus J.; Müller-Fielitz H.; Raasch W.; Hoffmann A.; Schmidtke M.; Clement B. Zanamivir amidoxime- and N-hydroxyguanidine-based prodrug approaches to tackle poor oral bioavailability. J. Pharm. Sci. 2015, 104, 3208–3219. 10.1002/jps.24508. [DOI] [PubMed] [Google Scholar]
- Balannik V.; Wang J.; Ohigashi Y.; Jing X.; Magavern E.; Lamb R. A.; DeGrado W. F.; Pinto L. H. Design and pharmacological characterization of inhibitors of amantadine-resistant mutants of the M2 ion channel of influenza A virus. Biochemistry 2009, 48, 11872. 10.1021/bi9014488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires J. B. Calorimetry and thermodynamics in drug design. Annu. Rev. Biophys. 2008, 37, 135–151. 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- Salom D.; Hill B. R.; Lear J. D.; DeGrado W. F. pH-Dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry 2000, 39, 14160–14170. 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens J.; Klebe G. A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates. Angew. Chem., Int. Ed. 2006, 45, 985–989. 10.1002/anie.200502302. [DOI] [PubMed] [Google Scholar]
- Gkeka P.; Eleftheratos S.; Kolocouris A.; Cournia Z. Free energy calculations reveal the origin of binding preference for aminoadamantane blockers of influenza A/M2TM pore. J. Chem. Theory Comput. 2013, 9, 1272–1281. 10.1021/ct300899n. [DOI] [PubMed] [Google Scholar]
- Bennett C. H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976, 22, 245–268. 10.1016/0021-9991(76)90078-4. [DOI] [Google Scholar]
- Chipot C., Pohorille A., Eds. Free Energy Calculations. Theory and Applications in Chemistry and Biology; Springer Verlag: Berlin, NY, 2007. [Google Scholar]
- Cady S.; Wang T.; Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 Protein. J. Am. Chem. Soc. 2011, 133, 11572–11579. 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Ma C.; Fiorin G.; Carnevale V.; Wang T.; Hu F.; Lamb R. A.; Pinto L. H.; Hong M.; Klein M. L.; DeGrado W. F. Molecular dynamics simulation directed rational design of inhibitors targeting drug-resistant mutants of influenza A virus M2. J. Am. Chem. Soc. 2011, 133, 12834–12841. 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa B. J.; Choma N.; Brown S. Y.; Keigher N.; Garland W.; Fukuda E. K. Quantitation of the enantiomers of rimantadine in human plasma and urine by gas chromatography-mass spectrometry. J. Chromatogr., Biomed. Appl. 1988, 431, 343–352. 10.1016/S0378-4347(00)83102-3. [DOI] [PubMed] [Google Scholar]
- Choma N.; Davis P. P.; Edorn R. W.; Fukuda E. K. Quantitation of the enantiomers of rimantadine and its hydroxylated metabolites in human plasma by gas chromatography/mass spectrometry. Biomed. Chromatogr. 1992, 6, 12–15. 10.1002/bmc.1130060105. [DOI] [PubMed] [Google Scholar]
- Manchand P. S.; Cerutti R. L.; Martin J. A.; Hill C. H.; Merrett E. K.; Keech E.; Belshe R. B.; Connell E. V.; Sim I. S. Synthesis and antiviral activity of metabolite of rimantadine. J. Med. Chem. 1990, 33, 1992–1995. 10.1021/jm00169a029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.