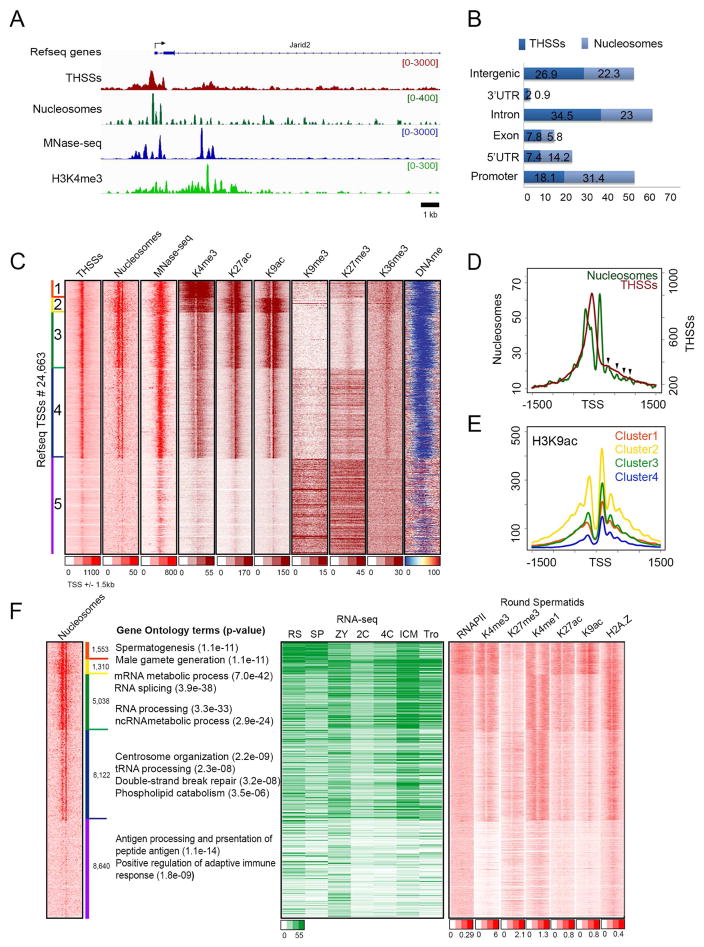
Figure 1. Epigenetic profiles of sperm chromatin at transcription start sites (TSS).
(A) Track view of THSS and nucleosome reads obtained using ATAC-seq, as well as ChIP-seq signal for H3K4me3 and RNAPII-Ser2ph around the Jarid2 TSS. MNase-seq data (Brykczynska, et al., 2010) is shown for comparison.
(B) Genome-wide distribution of THSSs and nucleosomes identified from sperm ATAC-seq. TSS ± 2 kb are listed as promoters.
(C) Heatmaps showing chromatin features around TSSs (± 1.5 kb). Sites are ordered by nucleosome and THSS signal from ATAC-seq.
(D) Average THSS (red) and nucleosome (green) ATAC-seq enrichment profiles relative to TSSs. Arrow heads indicate the position of nucleosomes downstream of the TSS.
(E) Average profiles of H3K9ac ChIP-seq for each cluster shown in panel C.
(F) Comparison of sperm nucleosome-occupied TSSs with gene ontology terms obtained using the GREAT tool, gene expression (RPKM from RNA-seq), and round spermatid chromatin state.
RS=round spermatid; SP=sperm; ZY=zygote; 2C= 2 cell embryo; 4C= 4 cell embryo; ICM= inner cell mass; Tro= trophectoderm.
See also Figures S1, S2 and S3