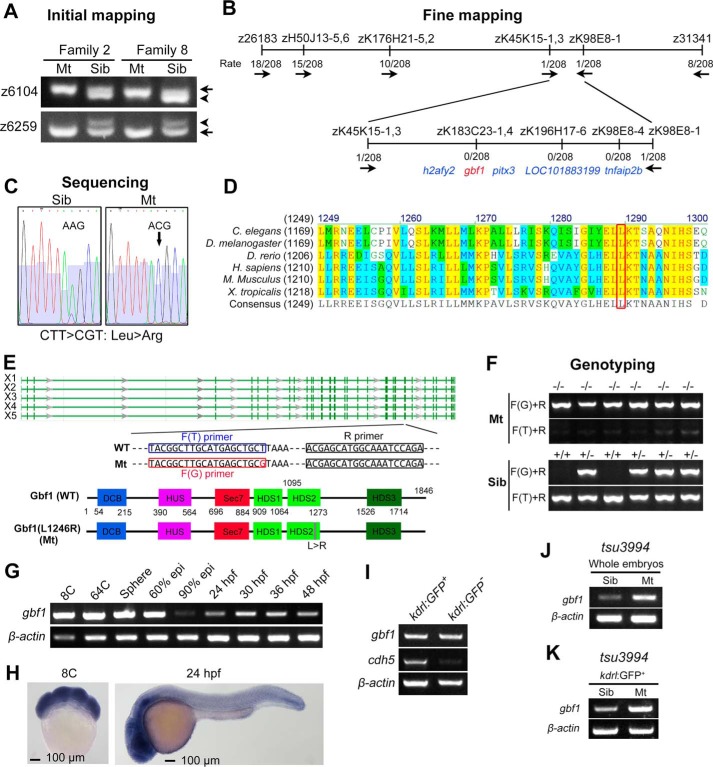
FIGURE 3.
Genetic mapping of mutant gene in tsu3994 mutants. A, initial mapping positioned the mutation site around the markers z6104 and z6259 on chromosome 13. Arrows and arrowheads indicate the polymorphic bands derived from Tu and India alleles, respectively. B, fine mapping results. The combination rates are shown, and the direction is indicated by arrows. C, a T → G point mutation was detected within the gbf1 coding sequence, presumably leading to Leu → Arg substitution in Gbf1 protein. D, the mutated leucine residue (boxed) of Gbf1 is evolutionally conserved across different species. E, genomic structure and splicing variants (X1–X5) of the zebrafish gbf1 locus and motif composition of Gbf1 protein with positions indicated. The sequences of primers used for genotyping were indicated. F, examples of genotyping results of mutants and siblings. G and H, expression of gbf1 transcripts in WT embryos at indicated stages was examined by RT-PCR (G) and in situ hybridization (H). I, transcription levels of gbf1 and cdh5 in kdrl:GFP+ ECs and in kdrl:GFP− non-ECs sorted from Tg(kdrl:GFP) embryos at 48 hpf were analyzed by RT-PCR. J and K, transcription levels of gbf1 in whole embryos (J) or in ECs sorted from tsu3994:Tg(kdrl:GFP) embryos (K) at 44 hpf were analyzed by RT-PCR.