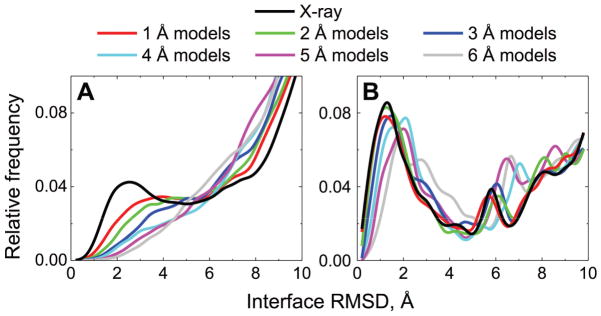
Figure 1. Distribution of near-native and false-positive matches according to the accuracy of protein models.
The top 1000 free docking (A) and all template-based docking (B) predictions, for each of the 165 complexes from the Models Docking Benchmark 2, at each of the six accuracy levels, were compared to the corresponding “ideal” complexes (see Methods) in terms of I-RMSD. In the docking of the X-ray structures, comparisons were made to the corresponding native X-ray structures. Near-native matches were defined as those with I-RMSD < 4 Å.