Abstract
Genetic diversity was assessed in the four orchid species using NGS based ddRAD sequencing data. The assembled nucleotide sequences (fastq) were deposited in the SRA archive of NCBI Database with accession number (SRP063543 for Dendrobium, SRP065790 for Geodorum, SRP072201 for Cymbidium and SRP072378 for Rhynchostylis). Total base pair read was 1.1 Mbp in case of Dendrobium sp., 553.3 Kbp for Geodorum sp., 1.6 Gbp for Cymbidium, and 1.4 Gbp for Rhynchostylis. Average GC% was 43.9 in Geodorum, 43.7% in Dendrobium, 41.2% in Cymbidium and 42.3% in Rhynchostylis. Four partial gene sequences were used in DnaSP5 program for nucleotide diversity and phylogenetic relationship determination (Ycf2 gene of Dendrobium, matK gene of Geodorum, psbD gene of Cymbidium and Ycf2 gene of Ryhnchostylis). Nucleotide diversity (per site) Pi (π) was 0.10560 in Dendrobium, 0.03586 in Geodorum, 0.01364 in Cymbidium and 0.011344 in Rhynchostylis. Neutrality test statistics showed the negative value in all the four orchid species (Tajima’s D value −2.17959 in Dendrobium, −2.01655 in Geodorum, −2.12362 in Rhynchostylis and −1.54222 in Cymbidium) indicating the purifying selection. Result for these gene sequences (matK and Ycf2 and psbD) indicate that they were not evolved neutrally, but signifying that selection might have played a role in evolution of these genes in these four groups of orchids. Phylogenetic relationship was analyzed by reconstructing dendrogram based on the matK, psbD and Ycf2 gene sequences using maximum likelihood method in MEGA6 program.
Keywords: Orchids, ddRADSeq, Genetic diversity, Phylogenetic analysis, Nucleotide diversity and evolution
Introduction
Orchids are famous for their ornamental and medicinal value (Swarts and Dixon 2009), belongs to the family Orchidaceae in the monocot. Orchids are traded as cut flowers in the international markets. Orchidaceae is a largest family in the flowering plant, which includes approximately 25,000 species (Atwood 1986; Hsu et al. 2011). Orchids can grow in the wide range of ecological habitats in the world and show maximum diversity among the species (Tremblay et al. 2005). Orchids grow in epiphytic and terrestrial habitat; they have acquired specialized reproductive and ecological strategies for advancing their adaptability in different environmental conditions. Flower of orchids form specialized structure by fusing androecium and gynoecium to form labellum and gynostegium for the smooth progression of pollination by attracting pollinators (birds and insects). This modification facilitates the co-evolution of orchid flowers and pollinators during the course of evolution (Yu and Goh 2001; Schiest et al. 2003). Orchids have been considered advance species among the flowering plants. It has been estimated that orchids originated at least 80–40 million years ago (Mya) (Dressler 1993) in the late Cretaceous eras. Some of the orchid species are considered rare and threatened because of their uncontrolled exploitation by the traders and orchids growers in addition to habitat deterioration due to modern industrial civilization (Swarts and Dixon 2009). Thus, protection measures are needed to conserve orchid germplasm of the world in the different ecological regions and habitat and thereby, the valuable gene pool, before these become extinct. Information on the genetic diversity in a gene pool of the species and their population structure is essential for conservation of any plant species to understand the growth pattern and survival potential of the germplasm (Pillon et al. 2007). Wide genetic diversity is the basic need to conserve any genetic resources (Brown 1988; Izawa et al. 2007), because it gives the adaptability features to the individual plant to cope with the environmental fluctuation in that ecogeographical habitat. If any individual is disappeared from the population due to any reason, it ultimately leads to loss of genetic traits (Izawa et al. 2007).
Genetic diversity helps to understand the evolutionary consequences in order to get idea about the genetic drift, mutational process and direction of gene-flow and divergence phenomena in the populations (Dressler 1993; Chase et al. 2005; George et al. 2009). Orchids contain high level of genomic complexity, because it carries wide range of variation in DNA content. DNA amount varies from 0.33 to 55.4 pg/cell, indicating the diversity in evolution (Leitch et al. 2009). Pattern of genetic diversity present in the organism indicates its complexity (Lynch and Conery 2003), and helps in sustainability of the ecosystem (Reusch et al. 2005). Species sensitivity to environmental changes is necessary for evolution (O’brien 1994). Less diversity in a population indicates the possibility for concealed endangerment (Amos and Balmford 2001). Genetic diversity was measured in orchid Calanthe tsoongiana for the conservation purpose of this endemic orchid in China (Qian et al. 2013).
Different molecular techniques were employed to measure the genetic diversity among plant species including some gene specific sequences such as matK, rbcL, ITS regions (Hilu and Liang 1997; Ma and Yin 2009; Cao et al. 2001; Koch et al. 2001; Zhu et al. 2003; Yang et al. 2004; Zhang et al. 2013; Chen et al. 2014; Xing 2014). Recently the high-throughput SNP genotyping techniques based on NGS method are used to analyze the genetic diversity (Kumar et al. 2012; Shirasawa et al. 2016). The NGS based advanced techniques such as genotyping-by-sequencing (GBS) (Elshire et al. 2011; Poland and Rife 2012) and restriction site-associated DNA sequencing (RAD-Seq) have become popular choice for genetic analysis because they are low cost and flexible (Davey et al. 2011) in experiment design.
To our knowledge, no published reports are available for nucleotide diversity in these four orchids species based on NGS technique. Therefore, in this study, we analyze preliminary data of the ddRAD sequencing report of four orchid species (Dendrobium densiflorum, Geodorum densiflorum, Cymbidium aloifolium and Rhynchostylis retusa) for evaluation of genetic diversity and evolutionary phylogenetic relationships based on DNA polymorphism using DnaSP5 and MEGA6 programs.
Materials and methods
Plant materials
Four orchid species were used in this study for DNA polymorphism analysis. Dendrobium densiflorum Lindl. and Geodorum densiflorum (Lam.) Schltr., Cymbidium aloifolium (L.) Sw. and Rhynchostylis retusa (L.) Blume were collected from Raiganj, Dist-Uttar Dinajpur, WB (Geographical location: Longitude-25.62°N and Latitude-88.12°E, Altitude- 40 m) and maintained in the garden of the Department of Botany, University of North Bengal, India.
DNA isolation and purification
Young leaves were harvested from the plants and kept in −20° C for 24 h before DNA extraction. Total genomic DNA was extracted using a Qiagen DNeasy plant mini kit (Qiagen, Germany). High quality DNA (concentration >300 ng/μl, A260/A280 ratio = 1.8–2.0 and A260/A230 ratio >1.7) was used for ddRAD sequencing purpose (Peterson et al. 2012).
Library construction for next generation sequencing (NGS)
Four DNA samples after meeting the required QC parameters were considered for library preparation using the standard protocols (Illumina) for next generation sequencing application. Following purification and quality check, about 1 μg genomic DNA of each of the four orchid species was double digested with SphI and MlucI restriction enzymes by incubating at 37 °C for 16–20 h following standard protocol (Peterson et al. 2012). The AMpure XP beads (Beckman Coulter Genomics) clean-up technology (with Dynabeads, Invitrogen) was used to clean the digested products (250–400 bp size) using standard protocols (Beckman Coulter Genomics) before library construction.
Adapters are ligated to the restricted cut sites in order to add barcodes and common PCR priming sequences. Barcodes are added to the downstream of the sequencing primer to resolve the products of different ligation after sequencing the library in NGS i.e., to separate the individual’s sequencing reads. Ligation of P1 (Barcoded) and P2 adapters was carried out using T4 DNA ligase followed by indexing with the addition of Index 1 and Index 2 sequences of 8 nt long. Adapter ligated fragments are separated through 2% agarose gel electrophoresis and visualized after staining with SYBR safe DNA stain. DNA fragments between 250 to 400 bp were excised from the gel under UV Trans illuminator. DNA fragments were extracted from the excised gel and purification was done by using AMpureBeads system. To increase the concentration of the DNA fragments of sequencing libraries, PCR amplification (8–12 cycles) was performed using Phusion™ polymerase kit. Products of PCR amplification were analyzed in Agilent Bioanalyzer to quantify molarity and fragment size distribution (BA profiles) in the sequencing library (NGS using Illumina). Each sequencing library was sequenced in six or more lanes using Illumina TruSeq chemistry on the HiSeq 2000 platform (SciGenom Labs Pvt. Ltd, Cochin, Kerala) and 90 bp paired-end reads were generated and used Solexapipeline version 1.0 to read the raw fluorescence images. To ensure high quality, raw data was filtered by deleting reads having adapter contamination or containing more than 50% low quality bases (quality value ≤ 5). The raw reads were assembled using SOAP de novo version 1.05 with default parameters (Peterson et al. 2012).
Analysis of nucleotide diversity from ddRAD sequencing data
Annotated and assembled contig sequences of ddRADSeq data were aligned using multiple sequence alignment software ClustlW (http://www.ebi.ac.uk/clustlw/). The output aligned files were saved as fasta format and used in an input file for further genetic and phylogenetic analysis in DnaSP5 and MEGA6. DNA polymorphism was studied by using DnaSP (Dna Sequence Polymorphism) program version 5.10.01 (Librado and Rozas 2009) (http://www.ub.es/dnasp/). DnaSP5 software was used to estimate the molecular diversity including the haplotype (gene) diversity (Hd), nucleotide diversity Pi (π) (Nei 1987), theta (θ) diversity (Watterson 1975), number of haplotypes (Nh), and number of segregating sites (S) in these four orchid species. Phylogenetic analysis was conducted using MEGA6 software program, and the phylogenetic tree generated using the Neighbor-Joining method (NJ) with 1000 bootstrap trials. The dendrograms were reconstructed based on partial gene sequences (matK, Ycf2, psbD) of four orchids. Pairwise synonymous (dS) and non-synonymous (dN) nucleotide substitution rates were estimated using the Jukes and Cantor (1969) distance model with the Nei–Gojobori method (Nei and Gojobori 1986) implemented in MEGA v6 (Tamura et al. 2013). The mean pairwise ratio of d N /d S (ω) was calculated and used to examine whether gene sequences evolve under purifying constraint for amino acid sequences (ω < 1) or positive selection for amino acid changes (ω > 1). Evidence for non-neutral evolution was investigated using the Tajima’s D (Tajima 1989), the Fu and Li’s D* and F* (Fu and Li 1993), the McDonald and Kreitman (1991), and the HKA (Hudson et al. 1987) tests, all implemented in the same DnaSP5 software.
Results and discussion
Analysis of nucleotide diversity
The raw sequence data (fastq format) of four orchid species (G. densiflorum, D. densiflorum, C. aloifolium and R. retusa) obtained from ddRAD sequencing technique are summarized in Table 1. Total sequence read was 553.3 Kbp in Geodorum (submitted to NCBI SRA archive with bioproject Accession no. SRP065790, PRJNA294125), 1.1 Mbp in Dendrobium (submitted to NCBI SRA archive with bioproject Accession no. SRP063543, PRJNA295128), 1.6 Gbp in Cymbidium (submitted to NCBI SRA archive with bioproject Accession no. SRP072201, PRJNA316048) and 1.4 Gpb in Rhynchostylis (submitted to NCBI SRA archive with bioproject Accession no. SRP072378, PRJNA316496). Total assembled transcripts were 285 in Geodorum with longest read length 275 bp, 14326 in Dendrobium with longest read length 512 bp, 126051 in Cymbidium with longest read length 423 bp and 42869 in Rhynchostylis with longest read length 484 bp. Present results of four orchids showed approximately same GC%, which was 43.9, 43.7, 41.2 and 42.3 in Geodorum, Dendrobium, Cymbidium and Rhynchostylis, respectively (Table 1). The GC content was 35.95% in case of Phalaenopsis equestris based on whole genome sequencing report (Jiangfeng et al. 2014) with genome size 1600 Mb (Diploid chromosome number 2n = 38, with 3.37 pg DNA/diploid genome). It indicated that P. equestris, has AT-rich genome with approximately 47,007 genes. In general, GC% in monocot plant ranges from 33.6 to 48.9% (Jiangfeng et al. 2014). The GC% in the four orchids species in the present investigation indicates that they have little higher GC% that signifies complex genome architecture compared to other plant groups. High GC% in the genome helps to adapt to the different environmental conditions (cold/dry seasons) leading to more complex gene regulation (Smarda et al. 2014). Partial genome sequence was carried out in one important orchid (Phalaenopsis equestris) (Hsu et al. 2011) and then whole genome was completely sequenced for its characterization and phylogenetic analysis (Cai et al. 2015). But reduced representation of the genomic segment has been sequenced in many organism using RAD, ddRAD, GBS methods based on NGS technology for genetic diversity and phylogenetic relationship using SNPs marker (Baird et al. 2008; Etter et al. 2011; Peterson et al. 2012; Jiangfeng et al. 2014; Wataru et al. 2014).
Table 1.
Summary of the ddRADSequencing contig assembly report (Illumina HiSeq 2000) along with NCBI SRA submission analysis and accession numbers
| ddRADSequencing report based on Illumina Hi-Seq 2000 platform | Orchid species | |||
|---|---|---|---|---|
| Geodorum densiflorum | Dendrobium densiflorum | Cymbidium aloifolium | Rhynchostylis retusa | |
| Total assembled transcripts | 285 | 14326 | 126051 | 42869 |
| Longest read length (bp) | 275 | 512 | 423 | 484 |
| Mean GC% | 43.9 | 43.7 | 41.2 | 42.3 |
| Read length ≥150 bp | 30 | 1695 | 19578 | 3791 |
| NCBI SRA accession no | SRP065790 (PRJNA2941 25) | SRP063543 (PRJNA295128) | SRP072201 PRJNA316048 | SRP072378 PRJNA316496 |
| SRA metadata spots | 7.0 k | 7.0 k | 7.9 M | 6.9 M |
| SRA bases | 553.3 kbp | 1.1 Mbp | 1.6 G bp | 1.4 Gbp |
| SRA size | 352.0 k | 617.7 k | 554.3 M | 482.0 M |
The Fastq files of four orchids were analyzed in the NCBI BLAST (http://blast.ncbi.nlm.nih.gov) with default parameters (such as plant, angiosperm, and monocot) to understand the taxonomic identity with the morphological identification. Blast analyses showed maximum similarity with the respective orchid species (Geodorum, Dendrobium, Cymbidium and Rhynchostylis) in the present investigation. Some of the contig assemblies from the orchid species were characterized and each of the annotated partial gene sequence was deposited in the NCBI GenBank with accession numbers (Table 2). DNA barcoding involves sequencing a standard region of DNA as a tool for species identification. Mainly the plastid DNA regions like matK and rbcL gene sequence data are used to identify species of land plants (Liu et al. 2013; Xu et al. 2015) and their phylogenetic relationship. Other gene sequences were also considered for evolutionary phylogenetic analysis purpose such as 28S, 18S, 16S rRNA gene, Ycf2 gene, GTP-transmembrane like sequences, NdhB2, Nad5, NADH dehydrogenase subunit-9, 30S ribosomal S11 like, D1 like photosystem II gene, PsbH protein like gene, Rps12 ribosomal gene like, tRNA-serine like gene, receptor protein like gene and many hypothetical protein genes. In the present investigation, partial gene sequences of matK gene (Geodorum densiflorum) (Accession no. KU136316, 235 bp) was used for nucleotide diversity analysis in DnaSP ver5 and also for phylogenetic relationship analysis in MEGA6.
Table 2.
Partial sequences of different genes characterized from the SRA data of ddRAD sequencing contig assembly for four orchids and submitted to NCBI GenBank
| GenBank accession no. | DNA length | Identified gene | GenBank accession no. | DNA length | Identified gene |
|---|---|---|---|---|---|
| Dendrobium densiflorum | |||||
| KU136279 | 280 bp | Ycf2 like gene | KU136294 | 249 bp | PsbH protein like gene |
| KU136279 | 367 bp | Hypothetical gene | KU136295 | 302 bp | Rps12 ribosomal gene |
| KU136281 | 222 bp | matK gene like | KU136296 | 466 bp | 18S rRNA like gene |
| KU136282 | 367 bp | Hypothetical gene | KU136297 | 512 bp | NdhH like gene |
| KU136283 | 277 bp | Ycf1 like gene | KU136298 | 271 bp | tRNA-Ser-like gene |
| KU136284 | 426 bp | D1 like Photo-II gene | KU136299 | 361 bp | 16S ribosomal protein |
| KU136285 | 289 bp | Ndh like gene | KU136300 | 390 bp | Hypothetical gene |
| KU136286 | 500 bp | Ycf4 like gene | KU136301 | 418 bp | Receptor protein like |
| KU136287 | 342 bp | Hypothetical gene | KU136302 | 426 bp | Nad5 like gene |
| KU136288 | 386 bp | Ycf2 like gene | KU136303 | 360 bp | Hypothetical gene |
| KU136289 | 426 bp | NADH dehy subunit-F | KU136304 | 329 bp | Hypothetical gene |
| KU136290 | 418 bp | Receptor protein gene | KU136305 | 304 bp | Hypothetical gene |
| KU136291 | 336 bp | NADH dehy subunit-9 | KU136306 | 304 bp | Hypothetical gene |
| KU136292 | 278 bp | Nad5 like gene | KU136307 | 500 bp | 30S ribosomal S11 like |
| KU136293 | 255 bp | 28S rRNA gene like | KU136308 | 385 bp | Hypothetical gene |
| Geodorum densiflorum | |||||
| KU136309 | 275 bp | NdhB2 like gene | KU136313 | 249 bp | matK like gene |
| KU136310 | 249 bp | GTP-trans like | KU136314 | 239 bp | Hypothetical gene |
| KU136311 | 245 bp | Ycf2 like | KU136315 | 234 bp | Hypothetical gene |
| KU136312 | 249 bp | matK like gene | KU136316 | 235 bp | matK like gene |
| Rhynchostylis retusa | |||||
| KX064256 | 363 bp | Ycf2 like | KX064258 | 484 bp | PsbD gene |
| KX064257 | 386 bp | SOC1 gene like | KX064259 | 425 bp | Rpl2 gene |
| Cymbidium aloifolium | |||||
| KX033492 | 484 bp | psbD gene | KX033493 | 296 bp | ATP synthase CF1 gene |
The matK is the chloroplast specific maturase K protein coding gene and widely used in phylogenetic relationship in plants. In case of Dendrobium densiflorum, partial gene sequence of Ycf2 (Accession no. KU136288, 386 bp) was used for nucleotide diversity analysis in DnaSP ver 5 and dendrogram reconstruction for phylogenetic analysis in MEGA6. Partial matK gene sequence of other orchids (13 in number) has been retrieved from NCBI GenBank by searching with matK gene sequence of Geodorum densiflorum of Raiganj for analysis purposes (Trillium discolor as out group).
The matK gene sequence of orchid Oncidium was used for phylogenetic relationship among 36 Oncidiinae species (Pan et al. 2012). In the same way partial gene sequence of Ycf2 for other orchids (10 in number) was retrieved from NCBI GenBank using Ycf2 sequence of Dendrobium densiflorum of Raiganj (Scadoxus cinnabarinus as out group). In case of Cymbidium aloifolium, partial gene sequence of psbD (Accession no. KX033492, 484 bp) was used for nucleotide diversity analysis in DnaSP ver 5 and dendrogram reconstruction for phylogenetic analysis in MEGA6. Partial psbD gene sequence of other orchids (8 in number) has been retrieved from NCBI GenBank by searching with psbD gene sequence of Cymbidium aloifolium of Raiganj for analysis purposes. In case of Rynchostylis, partial gene sequence of Ycf2 (Accession no. KX064256, 363 bp) was used for nucleotide diversity analysis in DnaSP ver 5 and dendrogram reconstruction for phylogenetic analysis in MEGA6. Partial Ycf2 gene sequence of other orchids (10 in number) has been retrieved from NCBI GenBank by searching with Ycf2 gene sequence of Rhynchostylis retusa of Raiganj for analysis purposes. Results of the DnaSP5 of four orchids were summarized in Table 3. Several plastid regions, such as matK, atpB, psbB, psbC and rpoC1, have been used to identify phylogenetic relationships in orchid (Cameron 2004; Cameron and Carmen Molina 2006) and results are consistent with our analysis in these four orchids also. To test the efficiency of gene sequences serving as markers in phylogenetic analyses, coding regions of four genes (accD, ccsA, matK, and ycf1) were used as a case study to construct phylogenetic trees in the subfamily Epidendroideae (including Dendrobium officinale Cypripedium macranthos) (Luo et al. 2014).
Table 3.
Nucleotide diversity and neutrality test reports obtained from DnaSP5 for four orchid species of Raiganj, India
| Parameter analysed in DnaSP5 | Geodorum densiflorum | Dendrobium densiflorum | Rhynchostylis retusa | Cymbidium aloifolium |
|---|---|---|---|---|
| Nucleotide sequences summary | ||||
| Number of sequence used | 14 | 11 | 10 | 08 |
| Number of sites | 243 | 415 | 363 | 485 |
| Excluding gaps/missing data | 232 | 344 | 277 | 398 |
| Number of polymorphic sites, S | 46 | 185 | 153 | 19 |
| Total number of mutation, Eta: η | 49 | 194 | 155 | 20 |
| Singleton variable sites | 40 | 179 | 150 | 17 |
| Parsimony informative sites | 6 | 6 | 3 | 2 |
| Haplotype/nucleotide diversity | ||||
| Number of haplotypes, h | 10 | 11 | 9 | 7 |
| Haplotype (gene) diversity, Hd | 0.945 | 1.00 | 0.9778 | 0.964 |
| Nucleotide diversity (per site), Pi: π | 0.03586 | 0.10560 | 0.11344 | 0.01364 |
| Average number of nucleotide differences, k | 8.31868 | 36.32727 | 31.42222 | 5.42857 |
| Theta (per sequence) from Eta | 15.40816 | 66.23493 | 54.79029 | 7.71350 |
| Theta (per site) from Eta | 0.06641 | 0.19254 | 0.19780 | 0.01938 |
| Neutrality tests | ||||
| Tajima’s D | −2.01655 | −2.17959 | −2.12362 | −1.54222 |
| Fu and Li’s D* test statistic | −2.63155 | −2.64773 | −2.49225 | −1.65366 |
| Fu and Li’s F* test statistic | −2.82891 | −2.87120 | −2.71298 | −1.81052 |
| Fu’s Fs statistic | −0.913 | −1.098 | 0.657 | −1.718 |
The haplotype (gene) diversity Hd was 1.00 and nucleotide diversity (per site) Pi (π) was 0.10560 in Dendrobium, which indicate that the genotype showing the genetic diversity. Nucleotide diversity (per site) Pi (π) was 0.03586 and haplotype (gene) diversity Hd was 0.945 in case of Geodorum while considering matK gene sequence. The matK sequence variation was low in the same group of orchids, but it was quite high in diverse groups. The value of Hd and Pi among orchid species (0.945 and 0.03586), respectively, signifies the high level of genetic diversity. Haplotype diversity (H d) was 0.945 in gene including 10 different haplotypes (in Geodorum sp.). On an average nucleotide diversities were π = 0.03586 and θ = 0.06641 in these 10 sequences of Geodorum. Both the value of nucleotide diversity, π and θ were high in this matK gene sequences. The value of θ was slightly higher than the value of π. Due to less SNP variation, the value of θ and π was similar and according to the neutrality test statistics it tends to be negative. The slipper orchids were analyzed to trace the phylogenetic relationship among the orchids species based on chloroplastic gene sequences (accD, rbcL, matK, rpoC1, rpoC2, ycf1, ycf2, and ndhF) in PAUP version 4.0b10 (Guo et al. 2012).
Pattern of molecular evolutionary mechanisms can be assessed using nucleotide variation present in the population. DNA sequence comparison can be the most important characteristics features to study the genetic variation in the natural population (Kreitman 1983). Natural selection played an important role in evolution (Ronald Fisher) and genetic drift has a minimum influence in evolution. But, Kimura (1968) has revolutionized the idea of evolution by proposing his Neutral theory of molecular evolution. According to the Neutral theory of molecular evolution, genetic changes that have occurred in the individual can spread by genetic drift. It can be detected by using the test (McDonald and Kreitman 1991). Tajima’s neutrality test (Tajima 1989) or D test statistic is used to test the Neutral theory of molecular evolution (Kimura 1983). Molecular differences found in the gene sequence, which arises through spontaneous mutation do not influence the fitness of the individual. In the present investigation of four orchid species, Tajima’s D test value was negative (−2.01655 in Geodorum, −2.17959 in Dendrobium, −2.12362 in Cymbidium and −1.54222 in Rhynchostylis), which suggests less polymorphisms in the population relative to expectation. This result also indicates the expansion of population size after a genetic bottleneck or a selective sweep (as a whole it is indicating purifying selection). A negative D value (Tajima’s D value) is usually interpreted as purifying selection. It was detected that the values for Tajima’s D, Fu and Li’s D* and F* were all statistically significant in the four orchids genus. Result for these gene sequences (matK and Ycf2 and psbD) indicate that they were not evolved neutrally signifying that selection might have played a role in evolution of these genes in these four groups of orchids.
Present investigation also showed the ratio of d N/d S is less than one (d N/d S < 1) indicating that they are mainly under negative selection or a recent population size expansion. Many factors are responsible for these selections, such as mutation, population size, recombination rate, gene conversion, and selection intensity (Hudson et al. 1987; McDonald and Kreitman 1991; Kreitman 2000). Tajima’s test or ‘D’ test statistic (Tajima 1989) was used to predict the probability of occurrence of Neutral theory of molecular evolution (Kimura 1983) in this present investigation. The present results are indicating the non neutral molecular evolution.
Results of the Tajima’s D test and Fu and Li’s D* and F* are listed in Table 3. These tests analyze the proportion of alleles at high or intermediate vs low frequencies among the four orchid species. Almost all the values are negative, indicating an excess of rare alleles in all the four orchids. An excess of rare alleles is consistent with positive selection, selective sweep or population size expansion. Present observation is fully consistent with the earlier reports in Mediterranean orchids (Aceto et al. 2007) for their genetic relationship based on gene sequences.
All the gene sequences (Ycf2 and matK, psbD) showed the bias in codon usage that prevailed in these orchids. Effective number of codon (ENC) was 51.00 in Dendrobium, 40.83 in Geodorum, 51.84 in Cymbidium, and 47.336 in Rhynchostylis. The codon bias index (CBI) was 0.543 in Dendrobium, 0.531 in Geodorum, 0.376 in Cymbidium, and 0.516 in Rhynchostylis indicating moderate codon bias. The CBI value indicates the deviation from the equal use of synonymous codon by the genes. This value ranges from 0 to 1 (0 = normal use of synonymous codon, 1 = high codon bias). Nonsynonymous rate variation among genes correlates most strongly with gene expression, perhaps owing to selection for translational robustness. Among lineages, perennial plants evolve more slowly than annuals, but the mechanism driving this effect remains unclear (Gaut et al. 2011). Evolutionary rates vary among nucleotide sites as a consequence of selection and mutational biases. These four orchids also showed the codon biases, which supports the earlier views (Gaut et al. 2011). Linkage disequilibrium (LD) values were measured in the four orchids genus by ZnS parameter (Kelly 1997), which were 0.6897 in case of Geodorum, 0.8630 in case of Dendrobium, 0.2175 in case of Cymbidium, and 0.9342 in Rhynchostylis (all are significant by the Bonferroni procedure).
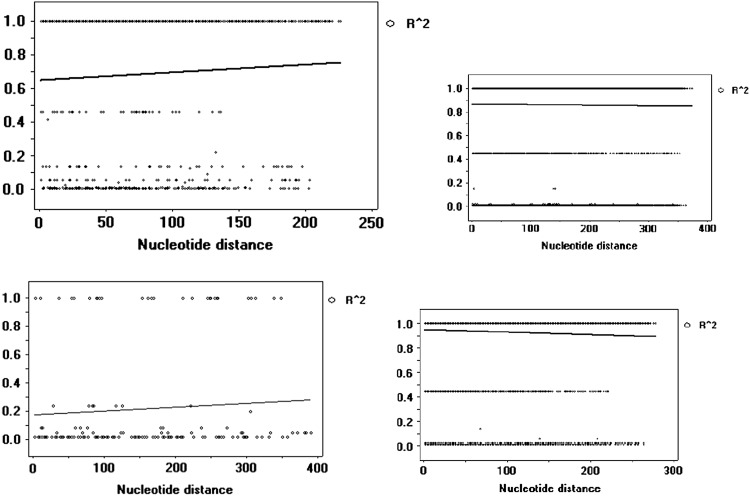
The ZnS value is the average of R2 value (Hill and Robertson 1968) over all sequences in pair wise comparison. If R2 value is = 1, it signifies that the two SNPs are in complete LD, which means if one can know one of the values then other value can be directly predicted. When R2 = 0, the two SNPs are considered to be independent. Graphical representation of the four orchid’s linkage disequilibrium was depicted in Fig. 1. Linkage Disequilibrium was significant for both G. densiflorum (R2 > 0.6897) and D. densiflorum (R2 > 0.8630) (0.001 < P < 0.01), R2 > 0.2175 in Cymbidium aloifolium and R2 > 0.9513 in Ryhnchostylis retusa. R2 values in the LD plots revealed slight decay of the analyzed sequences (Fig. 1). Pairwise comparisons showed significant value in Fisher’s exact test as well as in Chi-square test.
Fig. 1.
Linkage disequilibrium of Geodorum (from above left), Dendrobium, Cymbidium and Rhynchostylis depicted as graphical representation using matK, Ycf2 gene, and psbD gene respectively
The overall transition/transversion bias in matK gene sequences of Geodorum was R = 0.517, where R = [A*G*k 1 + T*C*k 2]/[(A + G)*(T + C)]. The analysis involved 14 nucleotide sequences with 232 positions in the final dataset (Table 4) and bold marked results for transitional substitutions and italics for transversionsal substitutions run in MEGA6 (Tamura et al. 2013). In case of Ycf2 gene sequences of Dendrobium, total bias for transition/transversion was R = 1.144 (Table 5). In case of psbD gene of Cymbidium, R = 1.023 using 8 gene sequences (Table 6). While considering the psbD gene sequence of Rhynchostylis retusa it showed transition/transversion bias rate R = 1.372 (Table 7). The emerging picture of the nucleotide substitution process in plants is a complex one. Evolutionary rates are seen to be quite variable, both among genes and among plant lineages. The present result (four orchids) is very much consistent with the views that the nucleotide substitution rates including transition/transversion ratio depends on genes and species. They are not evolving at the same level (Muse 2000).
Table 4.
Estimate of nucleotide substitution pattern based on MCL model in 14 matK gene sequences of Geodorum densiflorum of Raiganj
| A | T | C | G | |
|---|---|---|---|---|
| A | – | 12.12 | 4.98 | 0.88 |
| T | 9.84 | – | 9.9 | 4.71 |
| C | 9.84 | 24.09 | – | 4.71 |
| G | 1.84 | 12.12 | 4.98 | – |
Bold results are transitional substitutions and italics results are transversionsal substitutions based on MEGA6 analysis.
Table 5.
Nucleotide Substitution pattern was estimated using MCL model in 11 Ycf2 gene sequences of Dendrobium densiflorum of Raiganj
| A | T | C | G | |
|---|---|---|---|---|
| A | – | 6.82 | 4.99 | 10.1 |
| T | 7.01 | – | 10.96 | 3.77 |
| C | 7.01 | 14.97 | – | 3.77 |
| G | 18.8 | 6.82 | 4.99 | – |
Bold results are transitional substitutions and italics results are transversionsal substitutions based on MEGA6 analysis.
Table 6.
Estimate of nucleotide substitution pattern based on MCL model in 8 psbD gene sequences of Cymbidium aloifolium of Raiganj
| A | T | C | G | |
|---|---|---|---|---|
| A | – | 3.73 | 1.77 | 15.29 |
| T | 2.6 | – | 15.49 | 2.62 |
| C | 2.6 | 32.59 | – | 2.62 |
| G | 15.2 | 3.73 | 1.77 | – |
Bold results are transitional substitutions and italics results are transversionsal substitutions based on MEGA6 analysis.
Table 7.
Estimate of nucleotide substitution pattern based on MCL model in 10 Ycf2 gene sequences of Rhynchostylis retusa of Raiganj
| A | T | C | G | |
|---|---|---|---|---|
| A | – | 6.01 | 3.49 | 8.27 |
| T | 6.52 | – | 14.09 | 4.11 |
| C | 6.52 | 24.27 | – | 4.11 |
| G | 13.1 | 6.01 | 3.49 | – |
Bold results are transitional substitutions and italics results are transversionsal substitutions based on MEGA6 analysis.
Phylogenetic tree reconstructed in four orchids of Raiganj based of Ycf2, matK and psbD partial gene sequences
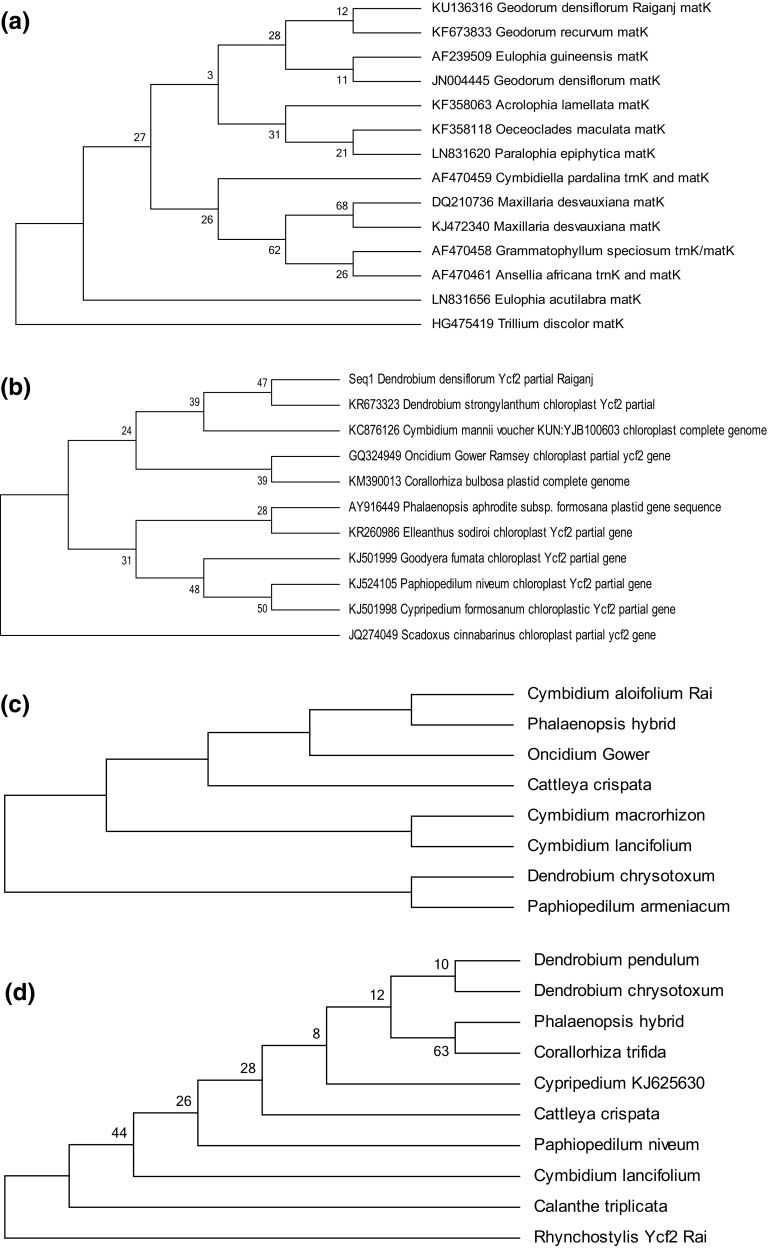
The evolutionary phylogenetic dendrograms were reconstructed depicting the genetic relatedness among the partial gene sequences (matK, Ycf2 and psbD) of four orchid species from Raiganj (Fig. 2a–d; Tables 8, 9, 10, 11) in MEGA6 (Tamura et al. 2013) (www.megasoftware.net/mega6.html). In case of Geodorum, matK gene sequences of 14 different orchids were analyzed based on pairwise distances between different gene sequences (Table 8). The maximum likelihood method was used to reconstruct the evolutionary history based on Jukes-Cantor model (Jukes and Cantor 1969). Bootstrap consensus tree was formed from 1000 replication to analyze the taxa (Felsenstein 1985). Heuristic search was employed to obtain initial trees by applying Neighbor-Join and BioNJ algorithms based on pairwise distance matrix. Gamma distribution was applied to trace the evolutionary rate differences among sites. Present analysis involved 14 nucleotide sequences in Geodorum (considering matK gene) 11 sequences in Dendrobium (considering Ycf2 gene), 8 in Cymbidium (considering psbD gene) and 10 in Rhynchostylis (considering Ycf2 gene). Three clusters were formed in the dendrogram of Geodorum. Geodorum densiflorum of Raiganj was closely related to the other Geodorum densiflorum (JN004445), and Geodorum recurvum (KF673833) species in this phylogenetic tree (Fig. 2a; Table 8). Trillium discolour was grouped as an out group and placed as separate clade in the dendrogram. The same method was applied to reconstruct the phylogenetic tree based on Ycf2 partial gene sequence of 11 species of Dendrobium (Fig. 2b; Table 9) and 10 sequences of Rhynchostylis (Fig. 2d; Table 15) based on pairwise genetic distances. Broadly, two clusters were formed in the dendrogram of Geodorum and closely related to other Geodorum species in this study (Fig. 2a) and Trillium discolor was used as out group, which was placed outside of the cluster. In case of Dendrobium densiflorum (Raiganj), two clusters were formed in the phylogenetic dendrogram and showed the close relationship with other Dendrobium species in this investigation (Fig. 2b), Scadoxus cinnabarinus was used as out group. In case of Rhynchostylis retusa, dendrogram formed one cluster and Rhynchostylis of Raiganj was placed in different clade which signifies that it was not closely related to other species considered in this dendrogram reconstruction (Fig. 2d; Table 11) using Ycf2 gene sequences. Three separate clusters were formed in the dendrogram while considering the psbD partial gene sequences of 8 orchid species including orchid Cymbidium of Raiganj (Fig. 2c; Table 10). One intriguing relationship that has been revealed through studies of branch lengths on molecular phylogenies is a link between the rate of molecular evolution and the net diversification rate. A correlation between evolutionary rates and species diversity has been found in several groups including flowering plants (Barraclough and Savolainen 2001). Present investigation showing different rates of phylogenetic diversification supporting the findings of other workers.
Fig. 2.
a Dendrogram reconstructed for molecular phylogenetic analysis using 14 matk gene partial sequences of orchid species by maximum likelihood method. b Dendrogram reconstructed for molecular phylogenetic analysis using 11 Ycf2 partial gene sequences of orchid species by maximum likelihood method. c Dendrogram reconstructed for molecular phylogenetic analysis using 8 psbD partial gene sequences of orchid species by maximum likelihood method. d Dendrogram reconstructed for molecular phylogenetic analysis using 10 Ycf2 partial gene sequences of orchid species by maximum likelihood method
Table 8.
Estimates of evolutionary divergence between sequences and pairwise distance among the 14 matK partial gene sequences of various orchids
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. KU136316 Geodorum densiflorum Raiganj matK | 0.009 | 0.004 | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 | 0.007 | 0.006 | 0.000 | 0.000 | 0.007 | 0.033 | |
| 2. LN831656 Eulophia acutilabra matK | 0.017 | 0.010 | 0.012 | 0.010 | 0.010 | 0.010 | 0.012 | 0.010 | 0.009 | 0.009 | 0.009 | 0.010 | 0.033 | |
| 3. AF239509 Eulophia guineensis matK | 0.004 | 0.022 | 0.007 | 0.009 | 0.009 | 0.009 | 0.007 | 0.009 | 0.008 | 0.004 | 0.004 | 0.009 | 0.034 | |
| 4. DQ210736 Maxillaria desvauxiana matK | 0.013 | 0.031 | 0.031 | 0.009 | 0.010 | 0.009 | 0.000 | 0.010 | 0.010 | 0.007 | 0.007 | 0.011 | 0.035 | |
| 5. AF470458 Grammatophyllum speciosum trnK/matK | 0.013 | 0.022 | 0.018 | 0.017 | 0.007 | 0.000 | 0.009 | 0.009 | 0.007 | 0.007 | 0.007 | 0.008 | 0.033 | |
| 6. AF470459 Cymbidiella pardalina trnK and matK | 0.013 | 0.022 | 0.018 | 0.022 | 0.013 | 0.007 | 0.010 | 0.006 | 0.004 | 0.007 | 0.007 | 0.006 | 0.032 | |
| 7. AF470461 Ansellia africana trnK and matK | 0.013 | 0.022 | 0.018 | 0.017 | 0.000 | 0.013 | 0.009 | 0.009 | 0.007 | 0.007 | 0.007 | 0.008 | 0.033 | |
| 8. KJ472340 Maxillaria desvauxiana matK | 0.013 | 0.031 | 0.013 | 0.000 | 0.017 | 0.022 | 0.017 | 0.010 | 0.010 | 0.007 | 0.007 | 0.011 | 0.035 | |
| 9. KF358118 Oeceoclades maculata matK | 0.013 | 0.022 | 0.018 | 0.026 | 0.017 | 0.009 | 0.017 | 0.026 | 0.004 | 0.007 | 0.007 | 0.006 | 0.032 | |
| 10. KF358063 Acrolophia lamellata matK | 0.009 | 0.017 | 0.013 | 0.022 | 0.013 | 0.004 | 0.013 | 0.022 | 0.004 | 0.006 | 0.006 | 0.004 | 0.031 | |
| 11. JN004445 Geodorum densiflorum matK | 0.000 | 0.017 | 0.004 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.009 | 0.000 | 0.007 | 0.033 | |
| 12. KF673833 Geodorum recurvum matK | 0.000 | 0.017 | 0.004 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.013 | 0.009 | 0.000 | 0.007 | 0.033 | |
| 13. LN831620 Paralophia epiphytica matK | 0.013 | 0.022 | 0.018 | 0.026 | 0.017 | 0.009 | 0.017 | 0.026 | 0.009 | 0.004 | 0.013 | 0.013 | 0.032 | |
| 14. HG475419 Trillium discolor matK | 0.201 | 0.200 | 0.205 | 0.218 | 0.199 | 0.195 | 0.199 | 0.218 | 0.194 | 0.189 | 0.201 | 0.201 | 0.195 |
Table 9.
Estimates of evolutionary divergence between sequences and pairwise distance among the 11 Ycf2 partial gene sequences of various orchids
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. Seq 1 Dendrobium densiflorum Ycf2 partial Raiganj | 0.100 | 0.008 | 0.007 | 0.007 | 0.007 | 0.007 | 0.006 | 0.005 | 0.006 | 0.003 | |
| 2. JQ274049 Scadoxus cinnabarinus chloroplast partial Ycf2 gene | 0.897 | 0.105 | 0.101 | 0.104 | 0.107 | 0.102 | 0.102 | 0.101 | 0.099 | 0.101 | |
| 3. KJ501999 Goodyera fumata chloroplast Ycf2 partial gene | 0.024 | 0.937 | 0.008 | 0.009 | 0.010 | 0.009 | 0.009 | 0.007 | 0.008 | 0.008 | |
| 4. KJ524105 Paphiopedilum niveum chloroplast Ycf2 partial gene | 0.018 | 0.901 | 0.024 | 0.007 | 0.008 | 0.009 | 0.007 | 0.004 | 0.006 | 0.006 | |
| 5. AY916449 Phalaenopsis aphrodite subsp. formosana plastid gene sequence | 0.015 | 0.915 | 0.027 | 0.015 | 0.007 | 0.008 | 0.006 | 0.005 | 0.005 | 0.006 | |
| 6. GQ324949 Oncidium Gower Ramsey chloroplast partial Ycf2 gene | 0.015 | 0.936 | 0.033 | 0.021 | 0.018 | 0.008 | 0.007 | 0.006 | 0.006 | 0.006 | |
| 7. KC876126 Cymbidium mannii voucher KUN:YJB100603 chloroplast complete genome | 0.015 | 0.908 | 0.033 | 0.027 | 0.024 | 0.024 | 0.008 | 0.007 | 0.008 | 0.006 | |
| 8. KM390013 Corallorhiza bulbosa plastid complete genome | 0.012 | 0.900 | 0.030 | 0.018 | 0.015 | 0.015 | 0.021 | 0.006 | 0.006 | 0.005 | |
| 9. KJ501998 Cypripedium formosanum chloroplastic Ycf2 partial gene | 0.012 | 0.902 | 0.018 | 0.006 | 0.009 | 0.015 | 0.021 | 0.012 | 0.004 | 0.005 | |
| 10. KR260986 Elleanthus sodiroi chloroplast Ycf2 partial gene | 0.012 | 0.891 | 0.024 | 0.012 | 0.009 | 0.015 | 0.021 | 0.012 | 0.006 | 0.005 | |
| 11. KR673323 Dendrobium strongylanthum chloroplast Ycf2 partial | 0.003 | 0.899 | 0.021 | 0.015 | 0.012 | 0.012 | 0.012 | 0.009 | 0.009 | 0.009 |
Table 10.
Estimates of evolutionary divergence between sequences and pairwise distance among the 8 psbD partial gene sequences of various orchids
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| 1. Cymbidium aloifolium Rai | 0.004 | 0.004 | 0.004 | 0.004 | 0.005 | 0.007 | 0.004 | |
| 2. Cymbidium macrorhizon | 0.008 | 0.004 | 0.004 | 0.004 | 0.006 | 0.007 | 0.000 | |
| 3. Cattleya crispata | 0.008 | 0.005 | 0.004 | 0.004 | 0.006 | 0.007 | 0.004 | |
| 4. Phalaenopsis hybrid | 0.005 | 0.008 | 0.008 | 0.004 | 0.005 | 0.007 | 0.004 | |
| 5. Dendrobium chrysotoxum | 0.008 | 0.005 | 0.005 | 0.008 | 0.006 | 0.006 | 0.004 | |
| 6. Oncidium Gower | 0.010 | 0.013 | 0.013 | 0.010 | 0.013 | 0.008 | 0.006 | |
| 7. Paphiopedilum armeniacum | 0.021 | 0.018 | 0.018 | 0.021 | 0.015 | 0.026 | 0.007 | |
| 8. Cymbidium lancifolium | 0.008 | 0.000 | 0.005 | 0.008 | 0.005 | 0.013 | 0.018 |
Table 11.
Estimates of evolutionary divergence between sequences and pairwise distance among the 10 Ycf2 partial gene sequences of various orchids
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. Rhynchostylis Ycf2 Rai | 0.157 | 0.164 | 0.167 | 0.156 | 0.156 | 0.175 | 0.157 | 0.139 | 0.156 | |
| 2. Cypripedium KJ625630 | 0.609 | 0.000 | 0.000 | 0.004 | 0.000 | 0.004 | 0.000 | 0.004 | 0.004 | |
| 3. Dendrobium pendulum | 0.617 | 0.000 | 0.000 | 0.004 | 0.000 | 0.004 | 0.000 | 0.004 | 0.004 | |
| 4. Dendrobium chrysotoxum | 0.620 | 0.000 | 0.000 | 0.004 | 0.000 | 0.004 | 0.000 | 0.004 | 0.004 | |
| 5. Paphiopedilum niveum | 0.596 | 0.004 | 0.004 | 0.004 | 0.004 | 0.005 | 0.004 | 0.005 | 0.005 | |
| 6. Phalaenopsis hybrid | 0.611 | 0.000 | 0.000 | 0.000 | 0.004 | 0.004 | 0.000 | 0.004 | 0.004 | |
| 7. Corallorhiza trifida | 0.639 | 0.004 | 0.004 | 0.004 | 0.007 | 0.004 | 0.004 | 0.005 | 0.005 | |
| 8. Cattleya crispata | 0.609 | 0.000 | 0.000 | 0.000 | 0.004 | 0.000 | 0.004 | 0.004 | 0.004 | |
| 9. Calanthe triplicata | 0.580 | 0.004 | 0.004 | 0.004 | 0.007 | 0.004 | 0.007 | 0.004 | 0.005 | |
| 10. Cymbidium lancifolium | 0.619 | 0.004 | 0.004 | 0.004 | 0.007 | 0.004 | 0.007 | 0.004 | 0.007 |
Table 15.
Codon based Z-test of neutral evolution using 10 nucleotide sequences of Ycf2 gene of Rhynchostylis sp.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. Rhynchostylis Ycf2 Rai | −0.595 | −0.595 | −0.841 | −0.353 | −0.595 | −0.458 | −0.595 | −0.353 | −0.626 | |
| 2. Cypripedium KJ625630 | 0.553 | 0.000 | −1.010 | −1.010 | 0.000 | 1.003 | 0.000 | −1.010 | −0.672 | |
| 3. Dendrobium pendulum | 0.553 | 1.000 | −1.010 | −1.010 | 0.000 | 1.003 | 0.000 | −1.010 | −0.672 | |
| 4. Dendrobium chrysotoxum | 0.402 | 0.315 | 0.315 | −1.442 | −1.010 | −0.667 | −1.010 | −1.442 | −1.194 | |
| 5. Paphiopedilum niveum | 0.725 | 0.315 | 0.315 | 0.152 | −1.010 | −0.667 | −1.010 | −1.442 | −1.194 | |
| 6. Phalaenopsis hybrid | 0.553 | 1.000 | 1.000 | 0.315 | 0.315 | 1.003 | 0.000 | −1.010 | −0.672 | |
| 7. Corallorhiza trifida | 0.647 | 0.318 | 0.318 | 0.506 | 0.506 | 0.318 | 1.003 | −0.667 | −0.358 | |
| 8. Cattleya crispata | 0.553 | 1.000 | 1.000 | 0.315 | 0.315 | 1.000 | 0.318 | −1.010 | −0.672 | |
| 9. Calanthe triplicata | 0.725 | 0.315 | 0.315 | 0.152 | 0.152 | 0.315 | 0.506 | 0.315 | −1.194 | |
| 10. Cymbidium lancifolium | 0.532 | 0.503 | 0.503 | 0.235 | 0.235 | 0.503 | 0.721 | 0.503 | 0.235 |
Comparative report of Tajima’s Neutrality Test (Tajima 1989) based on the partial gene sequences of matK, psbD and ycf2 was summarized in Table 16. The number of segregating sites was high (185) in ycf2 (in Dendrobium), low in psbD gene sequences (19) (in Cymbidium) but 153 in Rhynchostylis and 46 in Geodorum sp. Nucleotide diversity (π) was comparatively little high in case of Ycf2 sequences (0.105603) as compared to 0.011193 in Cymbidium psbD gene sequences and diversity was 0.035856 in matK sequences of Geodorum and 0.086563 in Rhynchostylis Ycf2 gene sequences.
Table 16.
Comparative summary report of Tajima’s neutrality test (Tajima 1989) based on matK, ycf2 and psbD partial gene sequences of four orchids of Raiganj
| Gene sequences | m | S | ps | Θ | π | D |
|---|---|---|---|---|---|---|
| Geodorum matK | 14 | 46 | 0.198276 | 0.062348 | 0.035856 | −1.858091 |
| Dendrobium Ycf2 | 11 | 185 | 0.537791 | 0.183611 | 0.105603 | −2.050025 |
| Rhynchostylis Ycf2 | 10 | 153 | 0.421488 | 0.148990 | 0.086563 | −2.086048 |
| Cymbidium psbD | 08 | 19 | 0.039175 | 0.015109 | 0.011193 | −1.345626 |
m = number of sequences, n = total number of sites, S = Number of segregating sites, p s = S/n, Θ = p s/a1, π = nucleotide diversity, D is the Tajima test statistic
The value of Tajima’s D statistic in both the cases was negative, suggesting that there may have occurred purifying selection (Table 16) in all four orchid species of Raiganj. Tajima’s test statistics is indicating noteworthy difference between the value of π and θ, thus, presenting the departure from the proposition of neutral theory. Neutrality test was conducted among these gene sequences using the codon-based Z-Test in MEGA6 software. The Nei-Gojobori method (Nei and Gojobori 1986) was employed to run this experiment. Null hypothesis rejection probability based on strict-neutrality equation (dN = dS), was depicted below diagonal in the following Tables 12, 13, 14, 15.
Table 12.
Codon based Z-test of neutral evolution using 14 nucleotide sequences of matK gene of Geodorum sp.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. KU136316 Geodorum densiflorum Raiganj matK | −0.144 | −1.011 | −0.422 | −1.222 | −0.417 | −1.222 | −0.422 | −0.417 | −0.701 | 0.000 | 0.000 | −0.415 | −1.569 | |
| 2. LN831656 Eulophia acutilabra matK | 0.886 | −0.797 | −0.407 | 0.095 | 2.270 | 0.095 | −0.407 | 2.270 | 2.024 | −0.144 | −0.144 | 2.270 | −1.190 | |
| 3. AF239509 Eulophia guineensis matK | 0.314 | 0.427 | −0.428 | −1.608 | −1.009 | −1.608 | −0.428 | −1.009 | −1.222 | −1.011 | −1.011 | −1.007 | −1.738 | |
| 4. DQ210736 Maxillaria desvauxiana matK | 0.673 | 0.685 | 0.669 | −1.608 | −0.805 | −1.608 | 0.000 | −0.607 | −0.802 | −0.422 | −0.422 | −0.604 | −1.590 | |
| 5. AF470458 Grammatophyllum speciosum trnK/matK | 0.224 | 0.924 | 0.111 | 0.110 | −0.425 | 0.000 | −1.608 | −0.162 | −0.422 | −1.222 | −1.222 | −0.158 | −1.051 | |
| 6. AF470459 Cymbidiella pardalina trnK and matK | 0.677 | 0.025 | 0.315 | 0.422 | 0.672 | −0.425 | −0.805 | 1.423 | 1.003 | −0.417 | −0.417 | 1.423 | −1.302 | |
| 7. AF470461 Ansellia africana trnK and matK | 0.224 | 0.924 | 0.111 | 0.110 | 1.000 | 0.672 | −1.608 | −0.162 | −0.422 | −1.222 | −1.222 | −0.158 | −1.051 | |
| 8. KJ472340 Maxillaria desvauxiana matK | 0.673 | 0.685 | 0.669 | 1.000 | 0.110 | 0.422 | 0.110 | −0.607 | −0.802 | −0.422 | −0.422 | −0.604 | −1.590 | |
| 9. KF358118 Oeceoclades maculata matK | 0.677 | 0.025 | 0.315 | 0.545 | 0.872 | 0.157 | 0.872 | 0.545 | 1.003 | −0.417 | −0.417 | 1.423 | −1.302 | |
| 10.KF358063 Acrolophia lamellata matK | 0.485 | 0.045 | 0.224 | 0.424 | 0.673 | 0.318 | 0.673 | 0.424 | 0.318 | −0.701 | −0.701 | 1.003 | −1.378 | |
| 11. JN004445 Geodorum densiflorum matK | 1.000 | 0.886 | 0.314 | 0.673 | 0.224 | 0.677 | 0.224 | 0.673 | 0.677 | 0.485 | 0.000 | −0.415 | −1.569 | |
| 12. KF673833 Geodorum recurvum matK | 1.000 | 0.886 | 0.314 | 0.673 | 0.224 | 0.677 | 0.224 | 0.673 | 0.677 | 0.485 | 1.000 | −0.415 | −1.569 | |
| 13. LN831620 Paralophia epiphytica matK | 0.679 | 0.025 | 0.316 | 0.547 | 0.875 | 0.157 | 0.875 | 0.547 | 0.157 | 0.318 | 0.679 | 0.679 | −1.290 | |
| 14. HG475419 Trillium discolor matK | 0.119 | 0.236 | 0.085 | 0.114 | 0.296 | 0.195 | 0.296 | 0.114 | 0.195 | 0.171 | 0.119 | 0.119 | 0.199 |
Table 13.
Codon based Z-test of neutral evolution using 11 nucleotide sequences of Ycf2 gene of Dendrobium sp.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. KU136288 Dendrobium densiflorum Ycf2 partial Raiganj | −0.740 | 0.740 | 0.485 | 2.016 | 2.259 | −0.036 | 1.002 | 1.743 | 1.743 | 1.002 | |
| 2. JQ274049 Scadoxus cinnabarinus chloroplast Ycf2 gene | 0.461 | −1.044 | −0.778 | −0.706 | −0.809 | −1.069 | −0.929 | −0.868 | −0.920 | −0.929 | |
| 3. KJ501999 Goodyera fumata chloroplast Ycf2 gene | 0.461 | 0.299 | −0.261 | 0.727 | 1.342 | 0.123 | 0.469 | −0.046 | 0.478 | 0.711 | |
| 4. KJ524105 Paphiopedilum niveum Ycf2 partial gene | 0.629 | 0.438 | 0.795 | 0.474 | 1.121 | −0.088 | 0.204 | −0.346 | 0.212 | 0.459 | |
| 5. AY916449 Phalaenopsis aphrodite plastid gene sequence | 0.046 | 0.482 | 0.468 | 0.636 | 2.682 | 0.480 | 1.742 | 1.742 | 1.742 | 2.015 | |
| 6. GQ324949 Oncidium chloroplast partial Ycf2 gene | 0.026 | 0.420 | 0.182 | 0.265 | 0.008 | 0.694 | 2.015 | 2.478 | 2.478 | 2.258 | |
| 7. KC876126 Cymbidium mannii chloroplast complete genome | 0.972 | 0.287 | 0.902 | 0.930 | 0.632 | 0.489 | −0.347 | 0.216 | 0.218 | −0.055 | |
| 8. KM390013 Corallorhiza bulbosa plastid complete genome | 0.318 | 0.355 | 0.640 | 0.838 | 0.084 | 0.046 | 0.729 | 1.420 | 1.420 | 1.002 | |
| 9. KJ501998 Cypripedium formosanum Ycf2 partial gene | 0.084 | 0.387 | 0.963 | 0.730 | 0.084 | 0.015 | 0.829 | 0.158 | 1.420 | 1.742 | |
| 10. KR260986 Elleanthus sodiroi chloroplast Ycf2 gene | 0.084 | 0.359 | 0.633 | 0.832 | 0.084 | 0.015 | 0.828 | 0.158 | 0.158 | 1.742 | |
| 11. KR673323 Dendrobium strongylanthum Cp Ycf2 partial | 0.318 | 0.355 | 0.479 | 0.647 | 0.046 | 0.026 | 0.956 | 0.318 | 0.084 | 0.084 |
Table 14.
Codon based Z-Test of Neutral evolution using 8 nucleotide sequences of psbD gene of Cymbidium sp.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| 1. Cymbidium aloifolium Rai | −0.302 | −0.302 | 1.420 | −0.307 | 1.742 | 1.247 | −0.302 | |
| 2. Cymbidium macrorhizon | 0.763 | 1.420 | −0.303 | 1.420 | −0.010 | 2.872 | 0.000 | |
| 3. Cattleya crispata | 0.763 | 0.158 | −0.303 | 1.420 | −0.010 | 2.872 | 1.420 | |
| 4. Phalaenopsis hybrid | 0.158 | 0.762 | 0.762 | −0.308 | 1.742 | 1.246 | −0.303 | |
| 5. Dendrobium chrysotoxum | 0.759 | 0.158 | 0.158 | 0.759 | −0.016 | 2.681 | 1.420 | |
| 6. Oncidium Gower | 0.084 | 0.992 | 0.992 | 0.084 | 0.987 | 1.436 | −0.010 | |
| 7. Paphiopedilum armeniacum | 0.215 | 0.005 | 0.005 | 0.215 | 0.008 | 0.154 | 2.872 | |
| 8. Cymbidium lancifolium | 0.763 | 1.000 | 0.158 | 0.762 | 0.158 | 0.992 | 0.005 |
The value of dS means numbers of synonymous substitution and dN means numbers of nonsynonymous substitutions (per site basis). The Neutrality test statistics (dN − dS) have been shown above the diagonal in the respective tables. The P value less than 0.05 were considered significant (at the 5% level) and which were in italic. We found significant differences in the ratio of non-synonymous to synonymous substitutions between four orchid species differing in net diversification rate, and indicates the signal of population size changes or alteration in selection pressures that might be causing this relationship (Duchene and Bromham 2013) for molecular evolutionary forces (Table 16).
Conclusion
Based on the genetic diversity analysis using DnaSP5 software, it was found that the all four orchid species of Raiganj are carrying moderate level of genetic diversity compared with the same gene sequences of other orchid species. Genetic diversity was evaluated based on nucleotide diversity and haplotype (gene) diversity of four orchid species. According to the neutral test statistics, all four orchid species showed negative value in Tajima’s D test suggesting less variability in the natural populations of orchids, than expected. This may also indicate that population is under expansion after a genetic bottleneck and as a whole indicating the purifying selection. The ratio of dN/dS is less than one (dN/dS < 1) means population is under negative selection, which also reflects the population size expansion. Different types of factors are responsible for these selections, such as mutation, population size, recombination rate, gene conversion, and selection intensity. Thus these four orchids (Cymbidium, Dendrobium, Geodorum and Rhynchostylis) of Raiganj, can be conserved in their respective habitat (in situ conservation system) to evolve naturally in their eco-climatic conditions. Overall results illustrate the complex evolutionary pattern in the population of the four orchids under study. A variety of mechanisms could maintain variation of these loci at different levels such as point mutations, filtered by the action of a diffuse purifying selection and by different selective constraints acting on synonymous and non-synonymous sites.
References
- Aceto S, Montieri S, Sica M, Gaudio L. Molecular evolution of the OrcPI locus in natural populations of Mediterranean orchids. Gene. 2007;392:299–305. doi: 10.1016/j.gene.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Amos W, Balmford A. When does conservation genetics matter? Heredity. 2001;87:257–265. doi: 10.1046/j.1365-2540.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- Atwood JT. The size of Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana. 1986;9:16. [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough T, Savolainen V. Evolutionary rates and species diversity in flowering plants. Evolution. 2001;55:677–683. doi: 10.1554/0014-3820(2001)055[0677:ERASDI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Brown AHD. The genetic diversity of germplasm collections. In: Fraleigh B, editor. Proceeding of a workshop on the genetic evaluation of plant genetic resources. Toronto: Agriculture Canada; 1988. pp. 9–11. [Google Scholar]
- Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, Chen LJ, Xu Q, Bian C, Zheng Z, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol Phylogenet Evol. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Cameron KM, Carmen Molina M. Photosystem II gene sequences of psbB and psbC clarify the phylogenetic position of Vanilla (Vanilloideae, Orchidaceae) Cladistics. 2006;22:239–248. doi: 10.1111/j.1096-0031.2006.00102.x. [DOI] [Google Scholar]
- Cao H, Sasaki Y, Fushimi H, Komatsu K. Molecular analysis of medicinally-used Chinese and Japanese curcuma based on 18 s rRNA gene and trnK gene sequences. Biol Pharm Bull. 2001;24:1389–1394. doi: 10.1248/bpb.24.1389. [DOI] [PubMed] [Google Scholar]
- Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH. Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae) Ann Bot. 2005;95:191–199. doi: 10.1093/aob/mci012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-Y, Bao Z-X, Ying-Qu WL, Li ZZ. Genetic diversity and population structure of the medicinal orchid Gastrodia elata revealed by microsatellite analysis. Biochem Syst Ecol. 2014;54:182–189. doi: 10.1016/j.bse.2014.01.007. [DOI] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Dressler RL. Phylogeny and classification of the orchid family. Portland: Dioscorides Press; 1993. [Google Scholar]
- Duchene D, Bromham L. Rates of molecular evolution and diversification in plants: chloroplast substitution rates correlate with species-richness in the Proteaceae. BMC Evol Biol. 2013;13:65. doi: 10.1186/1471-2148-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter PD, Bassham S, Hohenlohe PA, Johnson EA, Creskow A. SNP discovery and genotyping for evolutionary genetics using RAD sequencing. Methods Mol Biol. 2011;772:157–178. doi: 10.1007/978-1-61779-228-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B, Yang L, Takuno S, Eguiarte LE. The patterns and causes of variation in plant nucleotide substitution rates. Annu Rev Ecol Evol Syst. 2011;42:245–266. doi: 10.1146/annurev-ecolsys-102710-145119. [DOI] [Google Scholar]
- George S, Sharma J, Yadon VL. Genetic diversity of the endangered and narrow endemic Piperia yadonii (Orchidaceae) assessed with ISSR polymorphisms. Am J Bot. 2009;96:2022–2030. doi: 10.3732/ajb.0800368. [DOI] [PubMed] [Google Scholar]
- Guo Y-Y, Luo Y-B, Liu Z-J, Wang X-Q. Evolution and biogeography of the slipper orchids: Eocene vicariance of the conduplicate genera in the old and new world tropics. PLoS ONE. 2012;7(6):e38788. doi: 10.1371/journal.pone.0038788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hilu K, Liang H. The matK gene: sequence variation and application in plant systematics. Am J Bot. 1997;6:830–839. doi: 10.2307/2445819. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Chung Y-L, Chen T-C, Lee Y-L, Kuo Y-T, Tsai W-C, Hsiao Y-Y, Chen Y-W, Wen-Luan W, Chen H-H. An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. Plant Biol. 2011;11:3. doi: 10.1186/1471-2229-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Kawahara T, Takahashi H. Genetic diversity of an endangered plant, Cypripedium macranthos var. rebunense (Orchidaceae): background genetic research for future conservation. Conserv Genet. 2007;8:1369–1376. doi: 10.1007/s10592-007-9287-1. [DOI] [Google Scholar]
- Jiangfeng H, Zhao X, Laroche A, Zhen-Xiang L, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2014 doi: 10.3389/fpls.2014.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot. 2001;3:534–544. doi: 10.2307/2657117. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Methods to detect selection in populations with application to human. Annu Rev Genomics Hum Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- Kumar S, Banks TW, Cloutier S. SNP discovery through next-generation sequencing and its applications. Int J Plant Genomics. 2012;20:831460. doi: 10.1155/2012/831460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Kahandawala I, Suda J, Hanson L, Ingrouille MJ, Chase MW, et al. Genome size diversity in orchids: consequences and evolution. Ann Bot. 2009;104:469–481. doi: 10.1093/aob/mcp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang K, Liu Z, Luo K, Chen S, Keli C. Identification of medical plants of 24 Ardisia species from China using the matKgenetic marker. Pharmacogn Mag. 2013;9:331–337. doi: 10.4103/0973-1296.117829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Hou B-W, Ding Xiao-Yu. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE. 2014;9(6):e99016. doi: 10.1371/journal.pone.0099016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Ma JM, Yin SH. Genetic diversity of Dendrobium fimbriatum (Orchidaceae), an endangered species, detected by inter-simple sequence repeat (ISSR) Acta Bot Yuan. 2009;31:35–41. [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Muse SV. Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol. 2000;42:25–43. doi: 10.1023/A:1006319803002. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- O’brien SJ. A role for molecular genetics in biological conservation. Proc Natl Acad Sci USA. 1994;91:5748–5755. doi: 10.1073/pnas.91.13.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan I-C, Liao D-C, Wu F-H, Daniell H, Singh ND, Chang C, et al. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical Oncidium breeding. PLoS ONE. 2012;7(4):e34738. doi: 10.1371/journal.pone.0034738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest ddRADSeq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoSOne. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon Y, Qamaruz-Zaman F, Fay MF, Hendoux F, Piquot Y. Genetic diversity and ecological differentiation in the endangered fen orchid (Liparis loeselii) Conserv Genet. 2007;8:177–184. doi: 10.1007/s10592-006-9160-7. [DOI] [Google Scholar]
- Poland JA, Rife TW. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome. 2012;5:92–102. doi: 10.3835/plantgenome2012.05.0005. [DOI] [Google Scholar]
- Qian X, Wang C-X, Tian M. Genetic diversity and population differentiation of Calanthe tsoongiana, a rare and endemic orchid in China. Int J Mol Sci. 2013;14:20399–20413. doi: 10.3390/ijms141020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch TBH, Ehlers A, Hammerli A, Worm B. Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Natl Acad Sci USA. 2005;102:2826–2831. doi: 10.1073/pnas.0500008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiest FP, Peakall R, Mant JG, Ibarra F, Schulz C, Franke S, Francke W. The chemistry of sexual deception in an orchid-wasp pollination system. Science. 2003;302:437–438. doi: 10.1126/science.1087835. [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Hirakawa H, Isobe S. Analytical workflow of double-digest restriction site-associated DNA sequencing based on empirical and in silico optimization in tomato. DNA Res. 2016 doi: 10.1093/dnares/dsw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smarda P, Bures P, Ettore P, Mucina L, Horova L, Leitch I. High GC content in genome. Proc Nat Acad Sci USA. 2014;111:E4096–E4102. doi: 10.1073/pnas.1321152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts ND, Dixon KW. Terrestrial orchid conservation in the age of extinction. Ann Bot. 2009;104:543–556. doi: 10.1093/aob/mcp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay RL, Ackerman JD, Zimmerman JK, Calvo RN. Variation in sexual reproduction in orchids and its evolutionary consequences: a spasmodic journey to diversification. Biol J Linn Soc. 2005;84:1–54. doi: 10.1111/j.1095-8312.2004.00400.x. [DOI] [Google Scholar]
- Wataru K, Kajuharu N, Atushi Fujiwara Y, Nakamura MY, Ojima N, et al. A ddRAD-based genetic map and its integration with the genome assembly of Japanese eel (Anguilla japonica) provides insight into genome evolution after the teleost-specific genome duplication. BMC Genom. 2014;15:233–237. doi: 10.1186/1471-2164-15-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Pop Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Xing Y. SNP array—a powerful platform to accelerate genetic studies and breeding. J Plant Biochem Physiol. 2014;2:e119. doi: 10.4172/2329-9029.1000e119. [DOI] [Google Scholar]
- Xu S, Li D, Huang L. Evaluation of the DNA barcodes in Dendrobium (Orchidaceae) from Mainland Asis. PLoS ONE. 2015;10:e0115168. doi: 10.1371/journal.pone.0115168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DY, Hirotoshi F, Cai SQ, Katsuko K. Molecular analysis of Rheum species used as Rhei Rhizoma based on the chloroplast matK gene sequence and its application for identification. Biol Pharm Bull. 2004;3:375–383. doi: 10.1248/bpb.27.375. [DOI] [PubMed] [Google Scholar]
- Yu H, Goh CJ. Molecular genetics of reproductive biology in orchids. Plant Physiol. 2001;127:1390–1393. doi: 10.1104/pp.010676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Yang Z, Wang Y, Hu Y, Song X, Xu C. Nucleotide polymorphism and haplotide diversity of RTCS gene in China elite Maize inbred lines. PLoS ONE. 2013 doi: 10.1371/journal.pone.0056495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Fushimi H, Cai SQ, Komatsu K. Phylogenetic relationship in the genus panax: inferred from chloroplast trnK gene and nuclear 18S rRNA gene sequences. Planta Med. 2003;69:647–653. doi: 10.1055/s-2003-41117. [DOI] [PubMed] [Google Scholar]