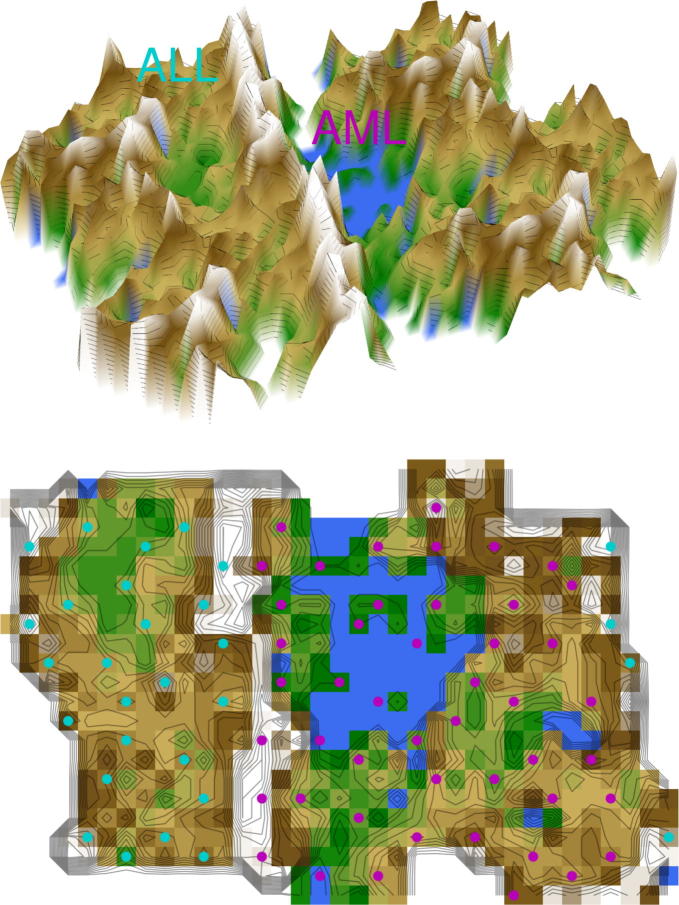
Fig. 5.
U-matrix representation of a classical dataset composed of microarray data of acute myeloid or lymphoblastic leukemia first used to propose the use of clustering algorithms to automatically discover distinctions between genetic profiles without previous knowledge of these classes [15] (data set #4). The figure shows the result of a projection of the data points onto a toroid grid 4000 neurons where opposite edges are connected. The cluster structure emerges from visualization of the distances between neurons in the high-dimensional space by means of a U-matrix [36]. The U-matrix was colored as a geographical map with brown or snow-covered heights and green valleys. Thus, valleys indicate clusters and watersheds indicate borderlines between different clusters. On the 3D-display (top) of the U-matrix, the valleys, ridges and basins can be seen. Valleys indicate clusters of similar drugs. The mountain range with “snow-covered” heights separates main clusters of leukemia. On the top view (bottom), the dots indicate the so-called “best matching units” (BMUs) of the self-organizing map (SOM), which are those neurons whose weight vector is most similar to the input. The BMUs are colored according to the obtained clustering of the data space, i.e., in magenta for acute myeloid leukemia (AML) and in light blue for acute lymphoblastic leukemia (ALL). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)