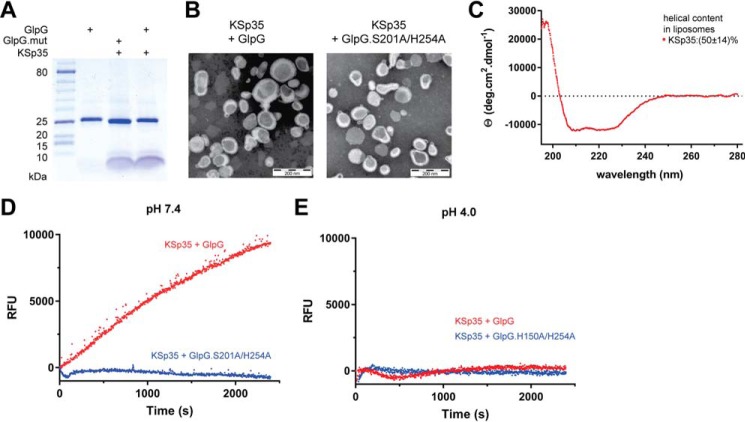
FIGURE 4.
The use of the transmembrane peptide substrate in liposomes. A, KSp35 was reconstituted into liposomes (LUVs) formed from E. coli polar lipid extract in the presence of GlpG or its inactive mutant S201A at pH 4.0. The resulting large unilamellar vesicles were analyzed by SDS-PAGE. B, the shape, lamellarity, and approximate size distribution of the KSp35+GlpG containing proteoliposomes formed at pH 4.0 were characterized by transmission electron microscopy. C, the integration of KSp35 into liposomes and its secondary structure content were analyzed by electronic CD. The substrate KSp35 (3 μm) was reconstituted with 2 mg/ml of E. coli polar lipid extract yielding an approximate peptide:lipid weight ratio of 1:500. D, activity of GlpG in liposomes detected by the KSp35 fluorogenic substrate. The substrate was co-reconstituted with wild type GlpG or its S201A/H254A mutant in a 30:1 molar ratio into LUVs made of E. coli polar lipid extract at pH 4.0, proteoliposomes were collected by ultracentrifugation and resuspended in 10 mm HEPES, 150 mm NaCl, pH 7.4, to start the cleavage reaction, which was then followed by measuring fluorescence at 493 nm. E, wild type GlpG or its H150A/H254A mutant were co-reconstituted with the substrate KSp35 in a 30:1 molar ratio into LUVs made of E. coli polar lipid extract at pH 4.0, proteoliposomes were collected by ultracentrifugation, resuspended in 50 mm sodium acetate, 150 mm NaCl, pH 4.0, and fluorescence was followed at 493 nm.